
Contribution of genetic polymorphism in ABCB1 to individual variations of imatinib plasma levels in patients with gastrointestinal stromal tumor
Highlight box
Key findings
• We conducted a comprehensive analysis of 1,485 blood samples from 192 patients with gastrointestinal stromal tumors (GISTs). Thirty-one single-nucleotide polymorphisms (SNPs) potentially associated with the pharmacogenetics of imatinib were genotyped with a SNP mass array platform. Polymorphism in ABCB1 rs1045642 was found to be significantly associated with steady-state imatinib trough plasma levels. The imatinib concentration of rs1045642 C carriers (CT + CC) was significantly higher than that in patients with the TT genotype, suggesting that a reduced dose of imatinib may be considered for allele C carriers of rs1045642 to reach an optimal response with tolerable side effects.
What is known and what is new?
• Imatinib mesylate has dramatically changed the treatment of GISTs, and several polymorphisms influencing the pharmacokinetic of imatinib have been identified.
• The ABCB1 rs1045642 genetic polymorphism may exert an effect on the pharmacokinetics of imatinib. The presence of the C-allele in ABCB1 rs1045642 is predictive for higher trough plasma concentrations of imatinib in patients with GIST.
What is the implication, and what should change now?
• For GIST patients with ABCB1 rs1045642 allele C, a decreased dose of imatinib might be considered to achieve high treatment response and low side effects.
Introduction
Gastrointestinal stromal tumors (GISTs) originate from the interstitial cells of Cajal and represent the most common mesenchymal malignancy of the gastrointestinal tract (1). Somatic mutations in the KIT or platelet-derived growth factor receptor alpha (PDGFRA) genes resulting in activation of the oncogenic tyrosine kinases play a crucial role in GIST tumorigenesis (2,3). Imatinib mesylate (IM), a multitargeted tyrosine kinase inhibitor (TKI), binds to the adenosine triphosphate (ATP)-binding pocket of the KIT/PDGFRA kinase domain, competitively inhibiting substrate phosphorylation and suppressing cellular proliferation (4). Since the advent of imatinib in 2002, the outcome of patients with GIST has been radically improved (5). IM has become the first-line therapy for patients with unresectable advanced GISTs, representing a milestone in targeted therapy for solid tumors. Therefore, adjuvant therapy with imatinib given at dose of 400 mg/day represent the standard treatment for patients with high risk of relapse after resection of the primary tumor.
However, clinical response varies significantly across individuals. Previous studies have shown that imatinib trough plasma concentrations (Cmin) can predict clinical prognosis in patients with GISTs. Patients with IM Cmin below 1,100 ng/mL experience a shorter time to progression and a lower rate of clinical benefit (6,7). In addition, despite it being a selective TKI, imatinib at a high concentration is associated with a broad range of toxicities, ranging from mild and amendable symptoms to rare but fatal hepatic failure (8,9). Thus, interindividual pharmacokinetic variations are important factors to be considered in the personalized treatment of GISTs.
Single-nucleotide polymorphisms (SNPs) are the most prevalent type of genetic polymorphisms and are known to potentially alter protein function (10). SNPs in genes involved in the pharmacokinetic pathways of imatinib may affect its efficacy and toxicity (11). This study aimed to investigate the relationship of genetic polymorphism, plasma imatinib levels, and clinical outcomes in GIST. A total of 1,485 blood samples from 192 consecutive patients with GIST were collected. A comprehensive panel of genetic polymorphisms related to the pharmacogenetics of IM was selected. SNP in the metabolizing genes (CYP3A4, CYP3A5, CYP2B6, POR, and NR1I2) and the transporter family genes (SLC22, SLC19, SLCO, and ABC) were genotyped with an SNP mass array platform. Angiogenesis-related genes (VEGFA, VEGFR2/KDR), cytokine receptor gene (IL4R), and epidermal growth factor receptor gene (EGFR) were also analyzed (12-14). We present this article in accordance with the REMARK reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-188/rc).
Methods
Study population
In this study, 192 Chinese patients with resected or advanced GIST receiving IM as adjuvant or first line therapy (Gleevec, Novartis Pharmaceuticals) at 400 mg daily were retrospectively enrolled between March 2009 and November 2020. All patients had histologically proven GISTs. Mutations of KIT and PDGFRA in the tumors were tested. The exclusion criteria were patients with severe comorbidities and those receiving drugs which could induce or inhibit CYP3A4 or P-glycoprotein. Patients with restricted oral administration were also excluded from the study. IM was regularly taken for at least 1 month to reach a steady state. A written informed consent was obtained from all patients before participation in the study. This study was performed in line with the principles of the Declaration of Helsinki (as revised in 2013) and approved by the Medical Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (ethical approval code: 2019-162).
IM trough plasma concentration determination
Peripheral blood was collected before the morning dose of IM, often 22–26 hours after the previous dose. The imatinib trough plasma concentrations (Cmin) were determined using a high-performance liquid chromatography (HPLC) method with UV-diode array detection (DAD), as described previously (15). The lower limit of quantification was 50 ng/mL. For each sample, at least three independent experiments were performed.
DNA extraction
Genomic DNA was extracted from whole blood using a DNA isolation kit (Qiagen, Hilden, Germany). A NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the concentration and purity of extracted DNA samples.
Genotyping
SNP genotyping was detected using a MassARRAY platform (Agena Bioscience, San Diego, CA, USA) (16). The forward, reverse, and single base extension primers are listed in Table S1. Targeted regions were amplified using multiplex polymerase chain reaction (PCR) as previously described (16). In each reaction, both positive and negative controls were included as quality control.
Statistical analysis
Statistical analyses were performed using SPSS version 21.0 (IBM, Armonk, NY, USA). A P value of 0.05 or less was considered statistically significant. GraphPad Prism (GraphPad Software, San Diego, CA, USA) was used to plot graphs. Mann-Whitney tests were used to analyze statistical differences of trough plasma concentrations between SNP genotypes, and the Kaplan-Meier method was used to compare progression-free survival (PFS) and overall survival (OS).
Results
Patients characteristics
The 192 patients who received IM at 400 mg daily were analyzed, and their demographic data and baseline clinical characteristics are summarized in Table 1. The mean age was 57.45 years (range, 23–83 years), and the number of males was 95 (49.48%). The average body weight was 60.07 kg (range, 40–90 kg), and the average body surface area (BSA) was 1.60 m2 (range, 0.85–2.05 m2). Mutational analysis was performed among the 170 patients. Mutations of the KIT or PDGFRA genes were detected in 150 (78.12%) patients, while in 20 patients (10.42%), no KIT or PDGFRA mutation was found. KIT mutations were detected in 148 patients (77.08%), mostly commonly in exon 11 (66.67%), followed by exon 9 (10.42%). Two patients (1.04%) had the PDGFRA mutation, both in exon 18. Moreover, 145 patients (75.52%) received IM as adjuvant treatment after surgery, and the remaining 47 patients (24.48%) received IM as palliative therapy for locally advanced and metastatic GISTs.
Table 1
| Characteristic | Value |
|---|---|
| Gender, female/male | 97 (50.52)/95 (49.48) |
| Age (years) | 57.45±11.55 [23–83] |
| Weight (kg) | 60.07±10.19 [40–90] |
| Height (cm) | 161.5±7.44 [145–176] |
| BSA (m2) | 1.60±0.17 [0.89–2.05] |
| Tumor site, stomach/other | 84 (43.75)/108 (56.25) |
| Maximum tumor diameter (cm) | 7.66±4.29 [1.3–25] |
| Mitotic index | |
| <5 | 91 (47.40) |
| 5–9 | 37 (19.27) |
| ≥10 | 25 (13.02) |
| Missing | 39 (20.31) |
| Primary mutation | |
| KIT exon 11 | 128 (66.67) |
| KIT exon 9 | 20 (10.42) |
| PDGFRA | 2 (1.04) |
| KIT/PDGFRA wild type | 20 (10.42) |
| Unknown | 22 (11.46) |
| Gastrectomy | |
| Total/partial | 73 (38.02) |
| None | 119 (61.98) |
| Indication for IM | |
| Adjuvant | 145 (75.52) |
| Palliative | 47 (24.48) |
Data are presented as n (%) or mean ± standard deviation [range]. BSA, body surface area; IM, imatinib mesylate.
IM steady-state trough plasma concentrations
A total of 1,485 blood samples were obtained from the 192 patients. The average follow-up time was 75.99 months, and the majority (97.40%) of patients underwent consecutive IM concentration tests at different time points, with a mean detection number of 7.73. The mean steady-state IM trough plasma concentration was 1,254.50 ng/mL, ranging from 548.81 to 2,162.30 ng/mL.
Effect of genetic polymorphisms on IM plasma levels
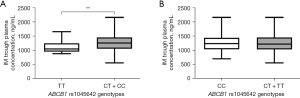
The comparisons of IM plasma trough levels across all genotypes are displayed in Table 2, Tables S2-S4, and Figure 1. For ATP-binding cassette subfamily B member 1 (ABCB1), the IM trough concentration (1,271.09±306.69 ng/mL) of rs1045642 C carriers (CT + CC) was significantly higher than of patients with the TT genotype (1,106.60±206.05 ng/mL) (P=0.008) (Figure 1, Table 2). In the other 30 selected SNPs, no significant differences were observed in IM plasma levels (Tables S2-S4). However, CYP3A4 rs2242480 CC genotype carriers tended to have higher imatinib trough concentrations (1,302.28±301.15 ng/mL) than did T carriers (CT + TT; 1,224.16±301.60 ng/mL) (P=0.07) (Table S2).
Table 2
| SNP_ID | Gene | Genotype | n | IM trough plasma concentration (ng/mL) | |
|---|---|---|---|---|---|
| Mean ± SD | P value | ||||
| rs1045642 | ABCB1 | CC | 80 | 1,258.07±307.25 | 0.84 |
| CT + TT | 112 | 1,249.55±298.40 | |||
| rs1045642 | ABCB1 | TT | 21 | 1,106.60±206.05 | 0.008** |
| CT + CC | 171 | 1,271.09±306.69 | |||
| rs2235040 | ABCB1 | GG | 177 | 1,261.66±309.71 | 0.62 |
| AG | 15 | 1,237.41±216.43 | |||
| rs1128503 | ABCB1 | TT | 70 | 1,256.11±321.83 | 0.64 |
| CT + CC | 122 | 1,253.86±290.32 | |||
| rs1128503 | ABCB1 | CC | 24 | 1,315.85±301.08 | 0.28 |
| CT + TT | 168 | 1,247.02±301.22 | |||
| rs12505410 | ABCG2 | GG | 18 | 1,250.94±330.37 | 0.68 |
| GT + TT | 174 | 1,253.33±299.42 | |||
| rs12505410 | ABCG2 | TT | 78 | 1,292.29±332.06 | 0.23 |
| GT + GG | 114 | 1,226.29±276.72 | |||
| rs2231142 | ABCG2 | CC | 104 | 1,260.36±290.86 | 0.91 |
| AC + AA | 88 | 1,259.09±318.83 | |||
| rs2231142 | ABCG2 | AA | 21 | 1,351.50±398.42 | 0.31 |
| AC + CC | 171 | 1,248.58±288.87 | |||
| rs2725250 | ABCG2 | AA | 98 | 1,293.19±326.92 | 0.11 |
| AG + GG | 94 | 1,225.31±273.92 | |||
| rs2725250 | ABCG2 | GG | 12 | 1,220.95±316.74 | 0.56 |
| AG + AA | 180 | 1,262.35±302.94 | |||
| rs2725252 | ABCG2 | AA | 40 | 1,294.78±315.85 | 0.21 |
| AC + CC | 152 | 1,247.21±297.11 | |||
| rs2725252 | ABCG2 | CC | 60 | 1,257.39±329.60 | 0.67 |
| AC + AA | 132 | 1,256.71±288.82 | |||
| rs9561765 | ABCC4 | GG | 101 | 1,251.49±293.31 | 0.69 |
| AG + AA | 91 | 1,269.07±315.14 | |||
| rs9561765 | ABCC4 | AA | 13 | 1,247.32±270.78 | 0.87 |
| AG + GG | 179 | 1,260.61±305.81 | |||
**, P<0.01. IM, imatinib mesylate; GIST, gastrointestinal stromal tumor; SD, standard deviation.
Treatment outcomes
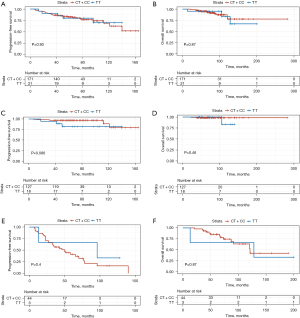
The mean PFS for the study population was 124.26 months [95% confidence interval (CI): 113.40–135.13] and the mean overall survival (OS) was 232.78 months (95% CI: 208.02–257.54). At the time of analysis, 43 patients (22.40%) progressed and 18 patients (9.38%) died. None of SNPs tested in this study showed a significant association with PFS or OS (Figure 2A,2B). Patients were further stratified into adjuvant or palliative groups according to whether the treatment administered involved resection of the primary tumor. Patients in palliative group received surgery because of emergency situations, like tumor rupture. There were no GISTs with tumor rupture during surgery. No association between GIST genotypes and clinical outcomes was observed in the adjuvant group (Figure 2C,2D) or palliative group (Figure 2E,2F).
Discussion
In the last 20 years, IM, which is the first small-molecule targeted drug with a known mechanism of efficacy, has dramatically improved the treatment of GISTs. However, a few problems still exist that restrict the clinical use of IM. One major challenge is the considerable interindividual variability in pharmacokinetics. The pharmacokinetic profile of IM depends on the proteins responsible for both the metabolism and transport of the compound. It was previously shown that the SNP of these variants may affect drug pharmacokinetics and antitumor therapy outcomes (17).
In recent years, numerous advancements have been made in this line of research, and several genetic polymorphisms affecting pharmacokinetics of imatinib have been identified. However, the current data are irreconcilable and not yet sufficiently conclusive to be translated into clinical use, which may be explained by several factors. One of these is that pharmacogenetic research is usually conducted on small-scale population cohorts with a limited number of time points during therapy, which may result in a lack of adequate statistical power and occasionally conflicting results. Therefore, we performed a comprehensive analysis with 1,485 blood samples from 192 patients with GIST. To the best of our knowledge, this is largest number of blood samples examined for a study of this kind. Another important and often overlooked aspect is the differences in detection methods used in SNP genotyping. Pyrosequencing has been widely used in previous research. However, a multicenter study demonstrated that compared with traditional pyrosequencing, MassARRAY, a SNP mass array platform and a robust tool involving multiplex PCR, has a higher accuracy for the detection of mutations (18). In our study, MassARRAY was used to ensure our findings were reliable and reproducible (19).
In this study, SNPs in the pharmacokinetic genes encoding for ABCB1, ABCG2, ABCC4, SLC22A1, SLC22A4, SLC22A5, SLCO1B3, SLC19A1, CYP3A4, CYP3A5, and CYP2B6 were analyzed. Several SNPs in relevant genes including VEGFA, VEGFR2/KDR, IL4R, and EGFR were also examined. ABCB1 is also known as multidrug resistance gene 1 (MDR1) and encodes for one of the main cellular membrane drug transporters (20). As an efflux pump, ABCB1, excretes a variety of endogenous and exogenous compounds including imatinib, thus affecting their blood concentrations (21). In this study, a significant difference was observed in steady-state IM trough plasma concentrations of patients with the ABCB1 (rs1045642) genotype. IM trough plasma levels in allele C carriers (CT + CC) were significantly higher than those in TT carriers (P=0.008), suggesting that the allele C of rs1045642 might be a significant factor for predicting the side effects commonly caused by high drug concentrations in the blood. Further research evaluating the functional significance of this polymorphism is warranted.
Notably, SNPs in the tested pharmacokinetic genes were not associated with a difference in PFS or OS, despite previous, sometimes conflicting, reports indicating otherwise (22-24). It is possible that most patients had IM trough concentrations higher than the threshold level required for clinical activity, negating any effects that these SNPs might have had on the actual serum concentrations above this level. Demetri et al. reported that when the imatinib plasma trough levels were below 1,100 ng/mL in patients with GIST, the PFS would show a significant decline (25). In our study, the IM plasma trough concentration was more than 1,100 ng/mL in most (67.19%) of the patients. IM were generally well tolerated and no severe adverse drug reactions were observed.
Conclusions
This pharmacogenetic study found the SNP in ABCB1 rs1045642 to be associated with increased trough plasma concentrations of IM in patients with GIST. A decreased dose of IM may be considered for allele C carriers of rs1045642 to achieve optimal response with tolerable side effects.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-188/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-188/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-188/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-188/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. A written informed consent was obtained from all patients before participation in the study. This study was performed in line with the principles of the Declaration of Helsinki (as revised in 2013) and approved by the Medical Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (ethical approval code: 2019-162).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Casali PG, Blay JY, Abecassis N, et al. Gastrointestinal stromal tumours: ESMO-EURACAN-GENTURIS Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2022;33:20-33. [Crossref] [PubMed]
- Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998;279:577-80. [Crossref] [PubMed]
- Masucci MT, Motti ML, Minopoli M, et al. Emerging Targeted Therapeutic Strategies to Overcome Imatinib Resistance of Gastrointestinal Stromal Tumors. Int J Mol Sci 2023;24:6026. [Crossref] [PubMed]
- Li GZ, Raut CP. Targeted therapy and personalized medicine in gastrointestinal stromal tumors: drug resistance, mechanisms, and treatment strategies. Onco Targets Ther 2019;12:5123-33. [Crossref] [PubMed]
- Mohammadi M, Gelderblom H. Systemic therapy of advanced/metastatic gastrointestinal stromal tumors: an update on progress beyond imatinib, sunitinib, and regorafenib. Expert Opin Investig Drugs 2021;30:143-52. [Crossref] [PubMed]
- Xia Y, Chen S, Luo M, et al. Correlations between imatinib plasma trough concentration and adverse reactions in Chinese patients with gastrointestinal stromal tumors. Cancer 2020;126:2054-61. [Crossref] [PubMed]
- Teranishi R, Takahashi T, Nishida T, et al. Plasma trough concentration of imatinib and its effect on therapeutic efficacy and adverse events in Japanese patients with GIST. Int J Clin Oncol 2023;28:680-687. [Crossref] [PubMed]
- Serrano C, George S. Gastrointestinal Stromal Tumor: Challenges and Opportunities for a New Decade. Clin Cancer Res 2020;26:5078-85. [Crossref] [PubMed]
- Hehlmann R. Chronic Myeloid Leukemia in 2020. Hemasphere 2020;4:e468. [Crossref] [PubMed]
- Jaruskova M, Curik N, Hercog R, et al. Genotypes of SLC22A4 and SLC22A5 regulatory loci are predictive of the response of chronic myeloid leukemia patients to imatinib treatment. J Exp Clin Cancer Res 2017;36:55. [Crossref] [PubMed]
- Omran MM, Abdelfattah R, Moussa HS, et al. Association of the Trough, Peak/Trough Ratio of Imatinib, Pyridine-N-Oxide Imatinib and ABCG2 SNPs 34 G>A and SLCO1B3 334 T>G With Imatinib Response in Egyptian Chronic Myeloid Leukemia Patients. Front Oncol 2020;10:1348. [Crossref] [PubMed]
- Blánquez-Martínez D, Díaz-Villamarín X, García-Rodríguez S, et al. Genetic Polymorphisms in VEGFR Coding Genes (FLT1/KDR) on Ranibizumab Response in High Myopia and Choroidal Neovascularization Patients. Pharmaceutics 2022;14:1555. [Crossref] [PubMed]
- Incorvaia L, De Biase D, Nannini M, et al. KIT/PDGFRA Variant Allele Frequency as Prognostic Factor in Gastrointestinal Stromal Tumors (GISTs): Results From a Multi-Institutional Cohort Study. Oncologist 2024;29:e141-51. [Crossref] [PubMed]
- Vincent CA, Nissen I, Dakhel S, et al. Epigenomic perturbation of novel EGFR enhancers reduces the proliferative and invasive capacity of glioblastoma and increases sensitivity to temozolomide. BMC Cancer 2023;23:945. [Crossref] [PubMed]
- Roth O, Spreux-Varoquaux O, Bouchet S, et al. Imatinib assay by HPLC with photodiode-array UV detection in plasma from patients with chronic myeloid leukemia: Comparison with LC-MS/MS. Clin Chim Acta 2010;411:140-6. [Crossref] [PubMed]
- Ma X, Zang X, Yang L, et al. Genetic polymorphisms in CYP2B6 may be associated with lung cancer risk in the Chinese Han population. Expert Rev Respir Med 2024; Epub ahead of print. [Crossref] [PubMed]
- Suttorp M, Bornhäuser M, Metzler M, et al. Pharmacology and pharmacokinetics of imatinib in pediatric patients. Expert Rev Clin Pharmacol 2018;11:219-31. [Crossref] [PubMed]
- Tsai SH, Chang PY, Wen YH, et al. Screening of single nucleotide polymorphisms within HLA region related to hematopoietic stem cell transplantation using MassARRAY technology. Sci Rep 2023;13:5913. [Crossref] [PubMed]
- Liu J, Xu Z, Li Y, et al. Comparison between MassARRAY and pyrosequencing for CYP2C19 and ABCB1 gene variants of clopidogrel efficiency genotyping. Mol Membr Biol 2019;35:1-8. [Crossref] [PubMed]
- Cerovska E, Salek C, Kundrat D, et al. ABC transporters are predictors of treatment failure in acute myeloid leukaemia. Biomed Pharmacother 2024;170:115930. [Crossref] [PubMed]
- Dalle Fratte C, Polesel J, Gagno S, et al. Impact of ABCG2 and ABCB1 Polymorphisms on Imatinib Plasmatic Exposure: An Original Work and Meta-Analysis. Int J Mol Sci 2023;24:3303. [Crossref] [PubMed]
- Verboom MC, Kloth JSL, Swen JJ, et al. Genetic polymorphisms in angiogenesis-related genes are associated with worse progression-free survival of patients with advanced gastrointestinal stromal tumours treated with imatinib. Eur J Cancer 2017;86:226-32. [Crossref] [PubMed]
- Jasek K, Grendar M, Stanclova A, et al. Prevalence and significance of M541L single nucleotide polymorphism in the central European cohort of gastrointestinal stromal tumor patients. J Cancer Res Clin Oncol 2021;147:1203-15. [Crossref] [PubMed]
- Unk M, Jezeršek Novaković B, Novaković S. Molecular Mechanisms of Gastrointestinal Stromal Tumors and Their Impact on Systemic Therapy Decision. Cancers (Basel) 2023;15:1498. [Crossref] [PubMed]
- Demetri GD, Wang Y, Wehrle E, et al. Imatinib plasma levels are correlated with clinical benefit in patients with unresectable/metastatic gastrointestinal stromal tumors. J Clin Oncol 2009;27:3141-7. [Crossref] [PubMed]


