
The highly expressed GOLPH3 in colorectal cancer cells activates smoothened to drive glycolysis and promote cancer cell growth and radiotherapy resistance
Highlight box
Key findings
• Golgi phosphoprotein 3 (GOLPH3) promoted colorectal cancer (CRC) progression and enhanced radiotherapy resistance via glycolysis mediated by the smoothened (SMO)-AMP-activated protein kinase (AMPK) axis.
What is known and what is new?
• GOLPH3 has been reported to affect the progression of various human cancers, including breast cancer, pancreatic ductal adenocarcinoma, non-small cell lung cancer, and hepatocellular carcinoma.
• Our study is the first of its kind to demonstrate the role of GOLPH3 in the radiotherapy resistance of CRC cells and to clarify the related molecular mechanisms.
What is the implication, and what should change now?
• Our findings indicated that GOLPH3 is a key regulator in CRC progression and the development of radiotherapy resistance. One of the mechanisms of the tumor-promoting effects of GOLPH3 might be its regulation of glycolysis as mediated by SMO-AMPK signaling. Further study based on clinic data are required to confirm the potential of GOLPH3 for use as a treatment target.
Introduction
As a frequently diagnosed cancer, colorectal cancer (CRC) accounted for approximately 550,000 new cancer cases and 286,000 deaths in China in 2020 (1). Over the past decade, CRC prevalence has declined in industrialized nations but has risen in China (2). In patients with CRC, the primary causes of treatment failure and death are distant metastasis and recurrence (1,3). Most patients with late-stage CRC are defined as initially unresectable, and the 5-year overall survival (OS) rate of these patients is less than 20% (4-6). Despite the significant progress made in refining molecular and immunological methods, the prognosis and treatment of advanced CRC remain unsatisfactory (7,8). In recent years, the standard treatment for CRC, especially local advanced rectal cancer, has included neoadjuvant radiotherapy (7,9). However, in clinical practice, treatment failure often occurs due to original or gradually adaptive radiotherapy resistance (10,11). Therefore, clarifying the molecular mechanisms related to radioresistance is critical for improving the prognosis of patients with CRC.
Golgi phosphoprotein 3 (GOLPH3) is located on the opposite sides of the Golgi apparatus, and its role involves protein glycation, protein classification, vesicle transport, and maintenance of the Golgi banding structure (12-14). Recently, GOLPH3 has been reported to affect the progression of various cancers. For example, in breast cancer (BC), GOLPH3 expression has been shown to be upregulated and its high expression associated with the poor prognosis (15). In pancreatic ductal adenocarcinoma (PDAC), GOLPH3 interacts with stress-induced phosphoprotein 1 and regulates telomerase activity to upregulate cell cycle-related proteins and facilitate tumor cell growth (16). In non-small cell lung cancer, GOLPH3 overexpression increases tumorigenicity and metastasis through activating Wnt/β-catenin pathway (17). In addition, GOLPH3 knockdown sensitizes lung adenocarcinoma cells to X-ray irradiation (18). The interaction between GOLPH3 and myosin-18A causes Golgi dispersal, impairing Golgi trafficking to enable cell survival after DNA damage (18). Gao et al. found that GOLPH3 promotes sorafenib resistance and angiogenesis in hepatocellular carcinoma (19). Yu et al. reported that oxaliplatin resistance of colon cancer cells could be reversed via the inhibition of GOLPH3, which may be related to the suppression of the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of the rapamycin (mTOR) pathway (20). However, whether GOLPH3 is involved in the radiotherapy resistance of CRC cells has not yet been fully determined.
In this study, we assessed GOLPH3 expression in terms of its prognostic value for CRC. Next, we examined the biological and radiosensitivity regulating effects of GOLPH3 on CRC cells by performing in vitro experiments. To further clarify the molecular mechanisms underlying GOLPH3’s role in CRC, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed based on the data from RNA sequencing (RNA-seq) and The Cancer Genome Atlas (TCGA) database. Finally, GOLPH3’s roles and the related molecular mechanisms were confirmed in vivo. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-193/rc).
Methods
Bioinformatics analysis
The messenger RNA (mRNA) expression data and related clinical data of CRC in TCGA were obtained from the UCSC Xena database (http://xena.ucsc.edu/). Differential expression analysis was performed using the R package “DESeq2” version 1.44.0 (The R Foundation of Statistical Computing, Vienna, Austria). The R package “clusterProfiler” version 4.12.2 was used for KEGG enrichment analysis. The graphical visualization was implemented via the R package “ggplot2” version 3.5.1. All statistical analyses were conducted using R software version 4.4.1. Statistical significance was defined as a P value less than 0.05. The differences between the two groups were compared using the Wilcoxon test. Survival analysis was performed through the Kaplan-Meier method. For correlation analysis, the Spearman correlation coefficient was calculated.
Human specimens
This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the Ethics Committee of Shandong Cancer Hospital & Institute Affiliated to Shandong First Medical University (No. SDTHEC 2023006022) and informed consent was taken from all the patients. CRC tissue and adjacent tissue from 41 patients were collected at Shandong Cancer Hospital & Institute Affiliated to Shandong First Medical University between July 2023 and June 2024. None of the patients underwent antitumor therapy, including immunotherapy, chemotherapy, targeted therapy, and surgery.
Cell culture
Human CRC cell lines (LoVo, SW480, HT-29, and HCT-116) were purchased from Nanjing Cobioer Biosciences Co., Ltd. (Nanjing, China). LoVo and SW480 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Sigma-Aldrich, St. Lous, MO, USA). HT-29 and HCT-116 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma-Aldrich) and McCoy’s 5a medium (Sigma-Aldrich), respectively. All media were mixed with 10% fetal bovine serum (FBS; Sigma-Aldrich). At 37 ℃, all cells were grown in an incubator with a 5% CO2 atmosphere.
Cell transfection
According to the manufacturer’s instructions, transfection was carried with Lipofectamine 3000 (Invitrogen, Themo Fisher Scientific, Waltham, MA, USA). HCT-116 cells were transfected with GOLPH3 overexpression vector (OE-GOLPH3) and negative control vector (OE-NC). SW480 cells were transfected with small interfering RNA (siRNA) targeting GOLPH3 (si1-GOLPH3, 5'-GTGGCTGTATGTTAATTGAAT-3'; si2-GOLPH3, 5'-GCATTGAGAGGAAGGTTACAA-3') and scrambled siRNA (si-NC, 5'-GTGTTAAATCTAGTGTGAGTT-3'). OE-GOLPH3, OE-NC, si1-GOLPH3, si2-GOLPH3, and si-NC were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
Radiosensitivity assay
HCT-116 and SW480 cells were subjected to different doses of X-ray (0, 2, 4, and 6 Gy) via a 6-MV linear accelerator (Elekta, Stockholm, Sweden). To assess GOLPH3’s effects on CRC radiotherapy resistance, HCT-116 and SW480 cells were subjected to ionizing radiation (IR) after transfection.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Through TRIzol (Thermo Fisher Scientific, Waltham, MA, USA), RNA was extracted from tissues and cells. Next, complement DNA (cDNA) was generated with a PrimeScriptRT reagent kit (Takara Bio, Kusatsu, Japan). Power SYBR Green PCR master mix (Applied Biosystems, Thermo Fisher Scientific) was employed for performing qRT-PCR. The quantification of GOLPH3 expression was calculated according to 2−ΔΔCT method. The primers’ sequence information is as follows: GOLPH3 forward, 5'-AAGGACCGCGAGGGTTACAC-3'; GOLPH3 reverse, 5'-CGTCTCATTCCACAAGCCTCT-3'; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5'-GAATGGGCAGCCGTTAGGAA-3'; and GAPDH reverse, 5'-CCCAATACGACCAAATCAGAGA-3'.
Western blotting
Proteins in cells and tissues were extracted using RIPA lysis buffer (Beyotime Biotechnology, Nantong, China). After being subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific). The membrane was incubated at 4 ℃ with primary antibodies acquired from Proteintech [GOLPH3, 19112-1-AP; GAPDH, 10494-1-AP; B cell lymphoma-2 (Bcl-2), 12789-1-AP; BCL2-associated X (Bax), 50599-2-Ig; cleaved (C)-caspase-3, 82707-13-RR; E-cadherin, 20874-1-AP; N-cadherin, 22018-1-AP; Vimentin, 80232-1-RR; AMP-activated protein kinase (AMPK), 10929-2-AP; phosphorylated (p)-AMPK, 83924-1-RR; pyruvate kinase M2 (PKM2), 15822-1-AP; Histone H3, 17168-1-AP; Rosemont, IL, USA], PTM Biolabs [H3 lysine 18 lactylation (H3K18la), PTM-1406RM; Chicago, IL, USA], and Abcam (Snail, ab216347; Cambridge, UK) after being blocked with 5% nonfat milk. The next day, a secondary antibody (ab7090; Abcam) was used to incubate the membrane for 1 hours. After being visualized by an enhanced chemiluminescence (ECL) kit (Thermo Fisher Scientific), protein bands were gauged via ImageJ software (US National Institutes of Health, Bethesda, MD, USA).
Cell Counting Kit-8 assay (CCK-8)
After being seeded in 96-well plates, HCT-116 and SW480 cells were cultured for 12, 24, 48, and 72 hours, respectively. Subsequently, cells were exposed to CCK-8 reagents (Dojindo, Kumamoto, Japan). Two hours later, at a wavelength of 450 nm, the optical density (OD) was tested with a microplate reader (Thermo Fisher Scientific).
Colony formation
HCT-116 and SW480 cells were seeded in six-well plates. Two weeks later, cells were fixed and stained in 4% paraformaldehyde (PFA; Solarbio, Beijing, China) and crystal violet (Sigma-Aldrich), respectively. Finally, the number of colonies with more than 50 cells was counted.
5-Ethynyl-2-deoxyuridine (EdU) assay
HCT-116 and SW480 cells were inoculated in 24-well plates. After being exposed to EdU assay (RiboBio, Guangzhou, China) for 2 hours, cells were fixed by 4% PFA and then treated with 0.5% Triton X-100 (Sigma-Aldrich). Subsequently, cell nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI; Beyotime Biotechnology). Under a fluorescence microscope (Olympus, Tokyo, Japan), we observed EdU-positive cells.
Flow cytometry
After indicated treatment, HCT-116 and SW480 cells were resuspended in Annexin V-FITC binding buffer (Beyotime Biotechnology). Annexin V-FITC and propidium iodide (PI) were used for incubating cells in the dark. Twenty minutes later, cell apoptosis was analyzed via flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Wound healing assay
HCT-116 and SW480 cells were inoculated into six-well plates and cultured in serum-free medium. A wound was created with a 200-µL sterile pipette tip when cell confluence reached 100%. Twenty-four hours later, the images of wound healing were recorded under a microscope (Leica, Wetzlar, Germany).
Transwell assay
HCT-116 and SW480 cell migration and invasion were assessed through the use of Transwell chambers (Costar-Corning, NY, USA). In brief, a serum-free cell suspension was added into the upper compartment of a migration (Matrigel-uncoated insert) or invasion (Matrigel-coated insert) chamber, and medium supplemented with 10% FBS was added into the lower chamber. After 24-hour culturing, the migrated and invaded cells were fixed and stained with 4% PFA and crystal violet, respectively. Finally, the number of migrated and invaded cells was recorded under a microscope.
Microarray analysis
RNA in SW480 cells with si-GOLPH3 or si-NC transfection was extracted for second-generation RNA-seq. Through TRIzol, total RNA was separated and purified. The amount and purity of total RNA were measured using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific), and the integrity of RNA was tested with the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). When the concentration was >50 ng/µL, RNA integrity number (RIN) value >7.0, and total RNA >1 µg, the subsequent experiments were performed. The mRNA with PolyA was specifically captured through two rounds of purification using oligo magnetic beads (Thermo Fisher Scientific). The obtained mRNA was fragmented using the NEBNextR Magnesium RNA Fragmentation Module (New England Biolabs, Ipswich, MA, USA) at 94 ℃ for 5–7 minutes. The fragmented RNA was synthesized into cDNA via SuperScript II Reverse Transcriptase (Invitrogen, Thermo Fisher Scientific). Next, Escherichia coli DNA polymerase I (New England Biolabs) and RNase H (New England Biolabs) were used for second strand synthesis, and then these complex double-strands of DNA and RNA were to converted into DNA double strands. Moreover, dUTP solution (Thermo Fisher Scientific) was incorporated into second strands. The blunt ends of double-stranded DNA were filled, and then an A base was added to each end to enable it to be connected to a linker with a T base at the end. The fragment size was screened and purified using magnetic beads. The second strand was digested with UDG enzyme (New England Biolabs), and then subjected to polymerase chain reaction (PCR) to form a library with a fragment size of 300±50 bp (strand-specific library). The PCR protocol was as follows: predenaturation at 95 ℃ for 3 minutes, denaturation at 98 ℃ for a total of 8 cycles of 15 seconds each, annealing at 60 ℃ for 15 seconds, extension at 72 ℃ for 30 seconds, and final extension at 72 ℃ for 5 minutes. Finally, we used the NovaseqTM 6000 (Illumina, San Diego, CA, USA) under the PE150 sequencing mode to perform double-end sequencing.
Measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR)
The changes of ECAR and OCR in HCT-116 and SW480 cells were assessed with the Seahorse XFp Metabolic Analyzer (Agilent). In brief, cells (1×105 cells per well) were cultured in a Seahorse XFp cell culture plate (Agilent) overnight. Before OCR measurement for 1 hour in a CO2-free incubator, cells were maintained in XF base medium containing 10-mM of glucose, 2-mM of glutamine, and 1-mM of pyruvate. Next, cells were incubated with 2-µM of oligomycin, 1-µM of carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and 0.5 of µM rotenone and antimycin A. For ECAR measurement, cells were maintained in XF base medium containing 2-mM of glutamine for 1 hour. Next, the cells were incubated with 10-mM of glucose, 1-µM of oligomycin, and 50-mM of 2-deoxyglucose (2DG).
Measurement of glucose uptake
Glucose uptake was determined via a glucose uptake colorimetric assay kit (ab136955; Abcam). Briefly, after glucose starvation, HCT-116 and SW480 cells were exposed to 2DG for 20 minutes, washed with phosphate buffered saline (PBS), and then lysed with extraction buffer. The lysates were incubated with neutralizing buffer. After centrifugation, the supernatants were incubated with reaction mix A. One hour later, the samples were extracted and heated at 90 ℃ for 40 minutes. After being cooled on ice, the samples were incubated with reaction mix B. At 412 nm, the absorbance was recorded by a microplate reader.
Measurement of lactate level
The lactate level was determined with the L-Lactate Assay Kit (ab65331; Abcam). Briefly, after the indicated treatment, cell culture supernatants were collected and then added with reaction mix. Thirty minutes later, at 450 nm, the absorbance was recorded with a microplate reader.
Chromatin immunoprecipitation (ChIP) assay
The Pierce Magnetic ChIP Kit (Thermo Fisher Scientific) was employed for completing ChIP assay. In brief, cells were exposed to 1% formaldehyde (Sigma-Aldrich) and then sonicated to obtain DNA fragments. Next, DNA fragments were incubated with anti-IgG (30000-0-AP; Proteintech) or anti-H3K18la-ChIP grade (PTM-1427RM; PTM Biolabs) at 4 ℃. The next day, DNA-protein immunocomplexes were obtained with protein A/G magnetic beads. After being eluted and purified, DNA samples were subjected to PCR.
Xenograft mice model
Animal experiments were performed under a project license (No. SDTHEC 2023006021) granted by the Animal Ethics Committee of Shandong Cancer Hospital & Institute Affiliated to Shandong First Medical University, in compliance with the guidelines of Shandong Cancer Hospital & Institute Affiliated to Shandong First Medical University for the care and use of animals. A protocol was prepared before the study without registration. Mice (GemPharmatech, Nanjing, China) in the sh-NC group (n=5) and sh-GOLPH3 group (n=5) were subcutaneously injected with SW480 cells with sh-NC or sh-GOLPH3 transfection. The xenograft tumor volume was measured every 3 days from the 10th day of injection. At the 31st day of injection, all mice were euthanized, and tumors were removed to be weighed and subjected to immunohistochemistry (IHC), immunofluorescence (IF), terminal-deoxynucleoitidyl transferase mediated nick end labeling (TUNEL) assay, and Western blotting.
IHC analysis
Tissues were embedded in paraffin after being fixed in 4% PFA and dehydrated in gradient alcohol. After being cut, tissue sections were exposed to 3% H2O2 and then blocked through application of 4% normal goat serum (Invitrogen). Before being incubated with secondary antibody (ab7090), sections were incubated at 4 ℃ overnight with primary antibodies (GOLPH3, 19112-1-AP; H3K18la, PTM-1406RM; Ki67, 84432-1-RR; Proteintech). After being stained with DAB (Beyotime Biotechnology), sections were counterstained with hematoxylin (Beyotime Biotechnology). Finally, under a microscope, sections were examined.
TUNEL staining
The One Step TUNEL Apoptosis Assay Kit (Beyotime Biotechnology) was applied for detecting cell apoptosis. In brief, tissue sections were incubated for 20 minutes with protease K without DNase. After being washed with PBS, sections were exposed to TUNEL reaction mixture for 60 minutes in the dark. DAPI was applied for counterstaining sections. Under a fluorescence microscope, TUNEL positive cells were surveyed.
IF staining
For tissue, sections underwent antigen retrieval via application of sodium citrate buffer. For cells, HCT-116 and SW480 cells were fixed with 4% PFA and permeabilized in Triton X-100. Before being incubated with primary antibodies, including smoothened (SMO; 20787-1-AP; Proteintech), PKM2 (15822-1-AP; Proteintech), and H3K18la (PTM-1406RM; PTM Biolabs), sections or cells were incubated with goat serum for 1 h. subsequently, sections were incubated with secondary antibodies (ab150077 or ab150080; Abcam) and then stained with DAPI. Finally, SMO expression was analyzed under a fluorescence microscope.
Statistical analysis
Statistical analyses were conducted with GraphPad Prism software (Dotmatics, Boston, MA, USA). Experiments were repeated three times independently. All data are displayed as the mean ± standard deviation. A P value less than 0.05 indicated statistical significance. The statistical significance was determined via a Student’s t-test and one-way analysis of variance (ANOVA).
Results
GOLPH3 expression was correlated with poor survival of patients with CRC
To investigate the correlation between GOLPH3 expression and CRC progression, Kaplan-Meier survival analysis was performed based on the data from TCGA database. Superior survival was observed in patients with CRC with low GOLPH3 expression (Figure 1A). Next, we assessed GOLPH3 expression in CRC tissues and their corresponding adjacent tissues. As displayed in Figure 1B-1D, GOLPH3 expression was markedly elevated in CRC tissues. Furthermore, GOLPH3 was overexpressed in CRC cell lines; and it was noticed that GOLPH3 was highly expressed in SW480 cells while weakly expressed in HCT-116 cells (Figure 1E).
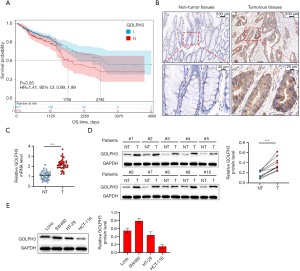
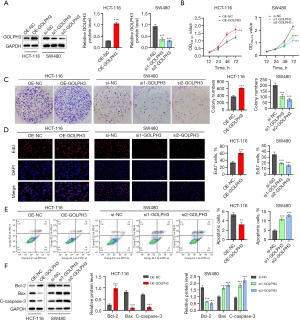
GOLPH3 promoted CRC cell proliferation and inhibited apoptosis
To further elucidate GOLPH3’s biological functions in CRC, GOLPH3 overexpression was induced in HCT-116 cells with lower GOLPH3 expression, and GOLPH3 was knocked down in SW480 cells with higher GOLPH3 expression (Figure 2A). As seen in Figure 2B, cell viability was promoted by GOLPH3 overexpression and inhibited by GOLPH3 knockdown. Additionally, GOLPH3 overexpression facilitated cell proliferation, but cell proliferation was suppressed by GOLPH3 knockdown (Figure 2C,2D). Flow cytometry revealed that cell apoptosis was reduced via GOLPH3 overexpression and increased by GOLPH3 knockdown (Figure 2E). Moreover, GOLPH3 overexpression increased Bcl-2 expression and decreased Bax and C-caspase-3 expression, but GOLPH3 knockdown produced the opposite effect (Figure 2F).
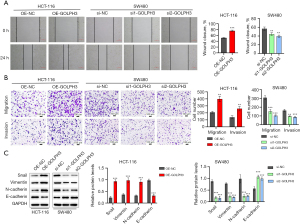
GOLPH3 promoted CRC cell migration and invasion
Similar to cell proliferation and apoptosis, CRC cell migration and invasion were also influenced by GOLPH3. Cell migration and invasion were promoted by GOLPH3 overexpression and inhibited by GOLPH3 knockdown (Figure 3A,3B). Furthermore, GOLPH3 overexpression in HCT-116 cells decreased E-cadherin expression and increased Snail, N-cadherin, and Vimentin expression, while the reverse was observed in SW480 cells with GOLPH3 knockdown (Figure 3C).
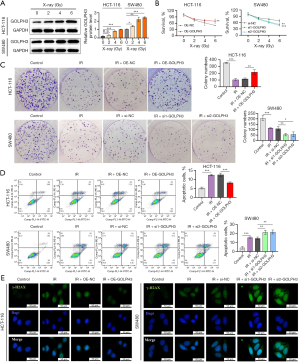
GOLPH3 promoted radiotherapy resistance of CRC cells
Next, we investigated GOLPH3’s change during radiotherapy for CRC. As shown in Figure 4A, X-rays increased GOLPH3 expression in a dose-dependent manner. As indicated by CCK-8 assay, colony formation assay, and flow cytometry, GOLPH3 overexpression promoted HCT-116 cell viability and proliferation and inhibited apoptosis under X-ray, but GOLPH3 knockdown enhanced radiosensitivity in SW480 cells (Figure 4B-4D). In addition, GOLPH3’s effects on DNA double-strand breaks (DSBs) were detected. GOLPH3 overexpression in HCT-116 cells reversed the increase of γ-H2AX induced by IR, while GOLPH3 knockdown in SW480 cells displayed the opposite effect (Figure 4E).
GOLPH3 promoted the membrane translocation of SMO in CRC cells
To further clarify the molecular mechanism underlying GOLPH3’s effect in CRC, RNA-seq was performed in SW480 cells with knockdown of GOLPH3. The differentially expressed genes (DEGs) between si-NC and si-GOLPH3 groups are shown in in Figure 5A, while the upregulated and downregulated genes are shown in Figure 5B. Next, we conducted KEGG enrichment analysis on these DEGs and found that these DEGs were enriched in many pathways, including Hedgehog signaling pathway (Figure 5C,5D). Based on TCGA database, KEGG enrichment analysis was also conducted. The results revealed that Hedgehog signaling pathway and glycolysis were regulated by GOLPH3 (Figure 5E,5F). A previous study indicated that SMO, a key protein in Hedgehog signaling pathway, could affect the glycolytic process in cells through regulating AMPK (21). In process of the infinite proliferation of cancer cells, glycolysis is a key pathological factor. Therefore, we speculated that SMO/AMPK pathway-mediated glycolysis might be the underlying mechanism involved in the oncogenic effect of GOLPH3. GOLPH3 is located on the Golgi apparatus and is mainly responsible for protein transport (22). SMO is mainly localized on the cell membrane. Through RNA-seq and TCGA analysis, we found that GOLPH3 affected Hedgehog signaling pathway and glycolysis but exerted no significant effect on the expression of SMO protein (Figure 5B). Therefore, we speculated that GOLPH3 might promote SMO’s membrane translocation and enhance its activity. As expected, IF indicated that GOLPH3 overexpression promoted the localization of SMO on the cell membrane, while GOLPH3 knockdown inhibited the localization of SMO on the cell membrane (Figure 5G).
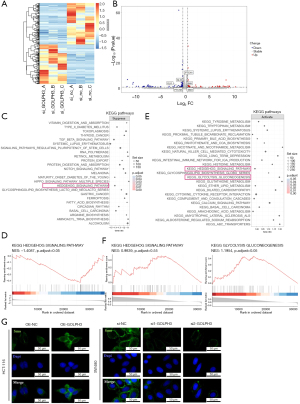
GOLPH3 promoted CRC cell glycolysis via activation of SMO/AMPK signaling
To verify our hypothesis that GOLPH3 could regulate CRC cell glycolysis, we examined CRC cell metabolism. GOLPH3 overexpression increased ECAR level and decreased OCR level in HCT-116 cells, while GOLPH3 knockdown decreased ECAR level and increased OCR level in SW480 cells (Figure 6A). Moreover, GOLPH3 overexpression enhanced glucose uptake and lactate production, while glucose uptake and lactate production were inhibited by GOLPH3 knockdown (Figure 6B,6C). The results of Western blotting revealed that GOLPH3 overexpression raised the levels of p-AMPK/AMPK, and PKM2, while GOLPH3 knockdown led to the opposite (Figure 6D). In addition, we used SMO agonist (SAG) and AMPK agonist (GSK621) to treat SW480 cells with GOLPH3 knockdown and found that the addition of SAG and GSK621 could reverse the inhibitory effect of GOLPH3 knockdown on glucose uptake and lactate production (Figure 6E).
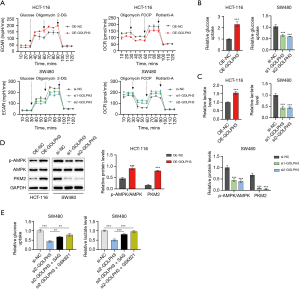
The lactate produced by CRC cells during glycolysis further increased GOLPH3 expression through H3K18la
An increasing number of studies have found that the glycolysis product lactate can cause protein lactylation modification, thereby affecting protein stability and expression (23-25), with H3K18la being the most intensely studied. Through Western blotting analysis, we found that H3K18la expression was increased in CRC tissues (Figure 7A), suggesting that H3K18 lactylation is common in CRC. Next, we sought to determine whether the lactylation of H3K18la affected GOLPH3 expression in HCT-116 and SW480 cells. We found that GOLPH3 expression increased with the increase in lactate concentration (Figure 7B). In contrast, glycolysis inhibitor (2DG) and LDHA inhibitor (oxamate) inhibited GOLPH3 expression in a concentration-dependent manner (Figure 7C,7D). Furthermore, oxamate inhibited H3K18la expression, but Nala (sodium lactate) reversed the inhibitory effect of oxamate on GOLPH3 and H3K18la expression (Figure 7E). In addition, the results of the ChIP and qPCR assay indicated that H3K18la was enriched on the GOLPH3 promoter, but 2DG and oxamate reduced the enrichment of H3K18la in the GOLPH3 promoter (Figure 7F,7G).
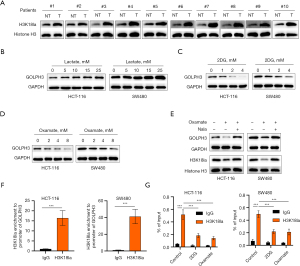
GOLPH3 knockdown inhibited the oncogenicity of CRC cells in vivo
To assess GOLPH3’s role in vivo, xenograft tumor experiments were conducted in mice. Mice in the sh-GOLPH3 group had smaller tumor volume and weight (Figure 8A-8C). Compared to the sh-NC condition, GOLPH3 knockdown reduced the number of Ki-67-positive cells and increased the proportion of apoptotic cells in tumor tissues (Figure 8D). Additionally, GOLPH3 knockdown decreased the abundance of SMO, PKM2, and H3K18la-positive cells in tumor tissues (Figure 8E,8F). Western blot analysis also confirmed that GOLPH3 knockdown inhibited GOLPH3, p-AMPK, PKM2, and H3K18la expression in tumor tissues (Figure 8G,8H).
Discussion
CRC has emerged as a worldwide health concern due to its high mortality (26,27). Therefore, the mechanism of CRC pathogenesis and novel targets for CRC treatment urgently need to be defined. As a phosphatidylinositol 4-phosphate (PI4P) binding protein, GOLPH3 is required for the regulation of Golgi glycosylation, vesicle trafficking, Golgi ribbon structure maintenance, and mitochondrial cardiolipin production (28,29). Recent studies have demonstrated that GOLPH3 expression is elevated in various cancers, such as BC, PDAC, prostate cancer, and glioma (15,16,30,31). In our study, we additionally found that GOLPH3 expression was increased in CRC tissues and cells and that a high expression of GOLPH3 was predictive of a poor prognosis. In vitro experiments suggested that GOLPH3 overexpression promoted CRC cell proliferation, migration, and invasion but inhibited apoptosis, as evidenced by the increase in Bcl-2, Snail, N-cadherin, and Vimentin expression and the decrease in Bax, C-caspase-3, and E-cadherin expression. Meanwhile, GOLPH3 knockdown reduced CRC cell proliferation, migration, and invasion but promoted apoptosis. Notably, GOLPH3 knockdown suppressed tumor growth in vivo. These findings suggest that GOLPH3 exerts carcinogenic effects within CRC.
In patients with locally advanced tumor, radiotherapy can provide effective tumor control (9,32). However, radioresistance response seriously threatens the quality of life of patients with CRC. In this study, we found that X-rays increased GOLPH3 expression in CRC cells. Furthermore, under IR, GOLPH3 overexpression promoted CRC cell proliferation and inhibited apoptosis, while GOLPH3 knockdown produced the opposite result. These results indicate that GOLPH3 enhances radiotherapy resistance in CRC cells. IR primarily eliminates tumor cells by inducing DNA damage, with DSBs being the most severe type of DNA damage (33). Tumor cells’ sensitivity to radiation largely depends on their ability to recognize and respond to DSBs (34). In our study, GOLPH3 overexpression decreased γ-H2AX (a sensitive DSB marker) expression under IR, while GOLPH3 knockdown further increased γ-H2AX expression. Taken together, these findings indicate that GOLPH3 promotes CRC cells’ radiotherapy resistance by inhibiting DSBs.
Metabolic reprogramming has been considered a hallmark of invasive cancer cells (35-37). Unlike normal cells, cancer cells prefer aerobic glycolysis even when oxygen is abundant (38). Glycolysis (also known as the Warburg effect) is characterized by a higher rate of glucose uptake and elevated production of adenosine triphosphate (ATP) and lactate, which promotes tumor progression (39,40). Recent studies have revealed that the inhibition of glycolysis represses CRC progression (41-43), which highlights the promising role of glycolysis as a target of therapy. In our study, KEGG enrichment analysis indicated that glycolysis could be regulated by GOLPH3. The in vitro experiments showed that GOLPH3 overexpression decreased OCR and increased ECAR, glucose uptake, and lactate production, while the opposite was observed in CRC cells with GOLPH3 knockdown. In addition, GOLPH3 knockdown inhibited PKM2 expression in cells and tumor tissues. In short, GOLPH3 promotes CRC progression through accelerating glycolysis.
To further clarify the complex mechanism underlying GOLPH3’s effect in CRC, we performed RNA-seq in CRC cells to identify the pathways affected by GOLPH3. Notably, the DEGs caused by GOLPH3 knockdown displayed enrichment in Hedgehog signaling pathway. Meanwhile, based on TCGA database, KEGG enrichment analysis also identified that Hedgehog signaling pathway was regulated by GOLPH3. As an evolutionarily conserved pathway, Hedgehog signaling is central to cell proliferation and differentiation, tissue development, and organ formation (44). Numerous studies have confirmed that the abnormal activation of Hedgehog signaling is related to the progression of multiple cancers, such as BC, endometrial cancer, and CRC (45-47). In Hedgehog signaling, transmembrane protein SMO is a key signal transducer (48). In the development of novel antitumor drugs, SMO inhibitors have emerged as an area of intense research in recent years (49). Teperino et al. reported that SMO could affect the glycolysis process in cells through regulating AMPK (21). In our study, GOLPH3 overexpression promoted the localization of SMO on the cell membrane and increased p-AMPK expression, while GOLPH3 knockdown led to opposite results. In addition, SMO and AMPK agonists reversed the inhibitory effect of GOLPH3 knockdown on glucose uptake and lactate production. In summary, we for the first time demonstrated that GOLPH3 activated the glycolysis of CRC cells through SMO/AMPK signaling. Further experiments including treating GOLPH3 knockdown cells with SAG and GSK621 to explore the mechanisms underlying the radiotherapy resistance of CRC are required in the following. Moreover, the regulation between GOLPH3 and glycolysis might be complex and multiple other signaling are involved. In addition to SMO/AMPK signaling, other signaling pathways involved in should be investigated further.
As a byproduct of glycolysis, lactate triggers histone lysine lactylation, thereby stimulating gene transcription (50). Recent studies have reported that histone lactylation, especially H3K18la, is involved in oncogenesis, tumor progression, tumor cellular metabolism reprogramming, and tumor immune escape (51-54). In our study, H3K18la expression was found to be increased in CRC tissues, suggesting that H3K18 lactylation is common in CRC. Furthermore, GOLPH3 expression was elevated by lactate treatment but reduced by 2DG and oxamate. Furthermore, the addition of Nala reversed the inhibitory effect of oxamate on GOLPH3 and H3K18la expression. In addition, ChIP and qPCR assay confirmed that H3K18la was enriched in the GOLPH3 promoter but that 2DG and oxamate partially reversed this effect. Overall, H3K18la mediated by lactate can be enriched on the GOLPH3 promoter, thereby promoting GOLPH3 expression.
Conclusions
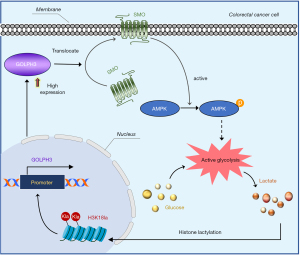
GOLPH3 was upregulated in CRC tissues, and low GOLPH3 expression was predictive of a better prognosis. Moreover, GOLPH3 knockdown inhibited malignant biological behaviors, reduced radiotherapy resistance, and impeded tumor growth in CRC. In terms of mechanism, GOLPH3 activated AMPK mediated glycolysis by promoting the localization of SMO on the cell membrane; meanwhile, lactate, the final product of glycolysis could induce H3K18 lactylation, and H3K18la was enriched in GOLPH3 the promoter, further increasing GOLPH3 expression (Figure 9). Our findings suggest that targeting the GOLPH3-SMO-AMPK axis has potential clinical value for CRC treatment. The feasibility of therapeutic strategies targeting GOLPH3 should be further validated in clinic.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-193/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-193/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-193/prf
Funding: This research was funded by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-193/coif). K.Z. reports funding from the Joint Fund for Tumor Prevention and Treatment of Shandong Natural Science Foundation (No. ZR2019LZL010). C.Z. reports funding from the Joint Fund for Innovation and Development of Shandong Natural Science Foundation (No. ZR2023LZL006). Z.G. reports funding from the National Natural Science Foundation of China (No. 82303347). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the Ethics Committee of Shandong Cancer Hospital & Institute Affiliated to Shandong First Medical University (No. SDTHEC 2023006022) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. SDTHEC 2023006021) granted by the Animal Ethics Committee of Shandong Cancer Hospital & Institute Affiliated to Shandong First Medical University, in compliance with the guidelines of Shandong Cancer Hospital & Institute Affiliated to Shandong First Medical University for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Wagle NS, Cercek A, et al. Colorectal cancer statistics, 2023. CA Cancer J Clin 2023;73:233-54. [Crossref] [PubMed]
- Li YJ, Wang X, Wu YJ, et al. Access to colorectal cancer screening in populations in China, 2020: A coverage-focused synthesis analysis. Int J Cancer 2024;155:558-68. [Crossref] [PubMed]
- Zeng H, Zhong X, Liu W, et al. Predicting treatment failure in stage III colon cancer patients after radical surgery. Front Oncol 2024;14:1397468. [Crossref] [PubMed]
- Tzeng CW, Aloia TA. Colorectal liver metastases. J Gastrointest Surg 2013;17:195-201; quiz p.201-2. [Crossref] [PubMed]
- Maeda Y, Shinohara T, Nagatsu A, et al. Long-Term Outcomes of Conversion Hepatectomy for Initially Unresectable Colorectal Liver Metastases. Ann Surg Oncol 2016;23:S242-8. [Crossref] [PubMed]
- Maughan TS, Adams RA, Smith CG, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet 2011;377:2103-14. [Crossref] [PubMed]
- Smith HG, Nilsson PJ, Shogan BD, et al. Neoadjuvant treatment of colorectal cancer: comprehensive review. BJS Open 2024;8:zrae038. [Crossref] [PubMed]
- Eng C, Yoshino T, Ruíz-García E, et al. Colorectal cancer. Lancet 2024;404:294-310. [Crossref] [PubMed]
- Kagawa Y, Smith JJ, Fokas E, et al. Future direction of total neoadjuvant therapy for locally advanced rectal cancer. Nat Rev Gastroenterol Hepatol 2024;21:444-55. [Crossref] [PubMed]
- Zhou Y, Shao Y, Hu W, et al. A novel long noncoding RNA SP100-AS1 induces radioresistance of colorectal cancer via sponging miR-622 and stabilizing ATG3. Cell Death Differ 2023;30:111-24. [Crossref] [PubMed]
- Akhunzianov AA, Rozhina EV, Filina YV, et al. Resistance to Radiotherapy in Cancer. Diseases 2025;13:22. [Crossref] [PubMed]
- Scott KL, Chin L. Signaling from the Golgi: mechanisms and models for Golgi phosphoprotein 3-mediated oncogenesis. Clin Cancer Res 2010;16:2229-34. [Crossref] [PubMed]
- Scott KL, Kabbarah O, Liang MC, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 2009;459:1085-90. [Crossref] [PubMed]
- Brauer BK, Chen Z, Beirow F, et al. GOLPH3 and GOLPH3L maintain Golgi localization of LYSET and a functional mannose 6-phosphate transport pathway. EMBO J 2024;43:6264-90. [Crossref] [PubMed]
- Song Q, Chen Q, Wang Q, et al. ATF-3/miR-590/GOLPH3 signaling pathway regulates proliferation of breast cancer. BMC Cancer 2018;18:255. [Crossref] [PubMed]
- Wang K, Jiang S, Huang A, et al. GOLPH3 Promotes Cancer Growth by Interacting With STIP1 and Regulating Telomerase Activity in Pancreatic Ductal Adenocarcinoma. Front Oncol 2020;10:575358. [Crossref] [PubMed]
- Song JW, Zhu J, Wu XX, et al. GOLPH3/CKAP4 promotes metastasis and tumorigenicity by enhancing the secretion of exosomal WNT3A in non-small-cell lung cancer. Cell Death Dis 2021;12:976. [Crossref] [PubMed]
- Chen G, Kong P, Yang M, et al. Golgi Phosphoprotein 3 Confers Radioresistance via Stabilizing EGFR in Lung Adenocarcinoma. Int J Radiat Oncol Biol Phys 2022;112:1216-28. [Crossref] [PubMed]
- Gao Y, Yin Z, Qi Y, et al. Golgi phosphoprotein 3 promotes angiogenesis and sorafenib resistance in hepatocellular carcinoma via upregulating exosomal miR-494-3p. Cancer Cell Int 2022;22:35. [Crossref] [PubMed]
- Yu T, An Q, Cao XL, et al. GOLPH3 inhibition reverses oxaliplatin resistance of colon cancer cells via suppression of PI3K/AKT/mTOR pathway. Life Sci 2020;260:118294. [Crossref] [PubMed]
- Teperino R, Amann S, Bayer M, et al. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell 2012;151:414-26. [Crossref] [PubMed]
- Li T, You H, Zhang J, et al. Study of GOLPH3: a potential stress-inducible protein from Golgi apparatus. Mol Neurobiol 2014;49:1449-59. [Crossref] [PubMed]
- Wang S, Huang T, Wu Q, et al. Lactate reprograms glioblastoma immunity through CBX3-regulated histone lactylation. J Clin Invest 2024;134:e176851. [Crossref] [PubMed]
- Zhang R, Li L, Yu J. Lactate-induced IGF1R protein lactylation promotes proliferation and metabolic reprogramming of lung cancer cells. Open Life Sci 2024;19:20220874. [Crossref] [PubMed]
- Tang F, Xiao D, Li X, et al. The roles of lactate and the interplay with m(6)A modification in diseases. Cell Biol Toxicol 2024;40:107. [Crossref] [PubMed]
- Santucci C, Mignozzi S, Malvezzi M, et al. European cancer mortality predictions for the year 2024 with focus on colorectal cancer. Ann Oncol 2024;35:308-16. [Crossref] [PubMed]
- Knudsen MD, Wang K, Wang L, et al. Colorectal Cancer Incidence and Mortality After Negative Colonoscopy Screening Results. JAMA Oncol 2025;11:46-54. [Crossref] [PubMed]
- Rizzo R, Parashuraman S, D'Angelo G, et al. GOLPH3 and oncogenesis: What is the molecular link? Tissue Cell 2017;49:170-4. [Crossref] [PubMed]
- Sechi S, Frappaolo A, Karimpour-Ghahnavieh A, et al. Oncogenic Roles of GOLPH3 in the Physiopathology of Cancer. Int J Mol Sci 2020;21:933. [Crossref] [PubMed]
- Kiełb P, Kaczorowski M, Kowalczyk K, et al. Comparative analysis of GOLPH3 expression in lymph node-positive prostate cancer: immunohistochemistry staining patterns and clinical significance. Front Oncol 2023;13:1265788. [Crossref] [PubMed]
- Wu S, Fu J, Dong Y, et al. GOLPH3 promotes glioma progression via facilitating JAK2-STAT3 pathway activation. J Neurooncol 2018;139:269-79. [Crossref] [PubMed]
- Fadlallah H, El Masri J, Fakhereddine H, et al. Colorectal cancer: Recent advances in management and treatment. World J Clin Oncol 2024;15:1136-56. [Crossref] [PubMed]
- Fujii S, Fuchs RP. Accidental Encounter of Repair Intermediates in Alkylated DNA May Lead to Double-Strand Breaks in Resting Cells. Int J Mol Sci 2024;25:8192. [Crossref] [PubMed]
- Ostoich PV. The Significance of the Response: Beyond the Mechanics of DNA Damage and Repair-Physiological, Genetic, and Systemic Aspects of Radiosensitivity in Higher Organisms. Int J Mol Sci 2024;26:257. [Crossref] [PubMed]
- Gao T, Yang L, Zhang Y, et al. Cancer metabolic reprogramming and precision medicine-current perspective. Front Pharmacol 2024;15:1450441. [Crossref] [PubMed]
- Mao Y, Xia Z, Xia W, et al. Metabolic reprogramming, sensing, and cancer therapy. Cell Rep 2024;43:115064. [Crossref] [PubMed]
- Aden D, Sureka N, Zaheer S, et al. Metabolic Reprogramming in Cancer: Implications for Immunosuppressive Microenvironment. Immunology 2025;174:30-72. [Crossref] [PubMed]
- Kooshan Z, Cárdenas-Piedra L, Clements J, et al. Glycolysis, the sweet appetite of the tumor microenvironment. Cancer Lett 2024;600:217156. [Crossref] [PubMed]
- Barba I, Carrillo-Bosch L, Seoane J. Targeting the Warburg Effect in Cancer: Where Do We Stand? Int J Mol Sci 2024;25:3142. [Crossref] [PubMed]
- Zhao J, Jin D, Huang M, et al. Glycolysis in the tumor microenvironment: a driver of cancer progression and a promising therapeutic target. Front Cell Dev Biol 2024;12:1416472. [Crossref] [PubMed]
- Chen J, Duan S, Wang Y, et al. MYG1 drives glycolysis and colorectal cancer development through nuclear-mitochondrial collaboration. Nat Commun 2024;15:4969. [Crossref] [PubMed]
- Liu H, Chen X, Wang P, et al. PRMT1-mediated PGK1 arginine methylation promotes colorectal cancer glycolysis and tumorigenesis. Cell Death Dis 2024;15:170. [Crossref] [PubMed]
- Zhang Y, Li L, Chu F, et al. Itraconazole inhibits tumor growth via CEBPB-mediated glycolysis in colorectal cancer. Cancer Sci 2024;115:1154-69. [Crossref] [PubMed]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001;15:3059-87. [Crossref] [PubMed]
- Liu R, Yu Y, Wang Q, et al. Interactions between hedgehog signaling pathway and the complex tumor microenvironment in breast cancer: current knowledge and therapeutic promises. Cell Commun Signal 2024;22:432. [Crossref] [PubMed]
- Yang H, Ji Y, Liu D, et al. LLGL2 targets the Hedgehog signaling pathway to influence malignant progression of endometrial cancer. Cell Signal 2025;127:111553. [Crossref] [PubMed]
- Chen J, Chen W, Li X, et al. CBC-1 as a Cynanbungeigenin C derivative inhibits the growth of colorectal cancer through targeting Hedgehog pathway component GLI 1. Steroids 2024;206:109421. [Crossref] [PubMed]
- Ogden SK, Fei DL, Schilling NS, et al. G protein Galphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature 2008;456:967-70. [Crossref] [PubMed]
- Rimkus TK, Carpenter RL, Qasem S, et al. Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers (Basel) 2016;8:22. [Crossref] [PubMed]
- Zhang D, Tang Z, Huang H, et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019;574:575-80. [Crossref] [PubMed]
- Yu J, Chai P, Xie M, et al. Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome Biol 2021;22:85. [Crossref] [PubMed]
- Yang J, Luo L, Zhao C, et al. A Positive Feedback Loop between Inactive VHL-Triggered Histone Lactylation and PDGFRβ Signaling Drives Clear Cell Renal Cell Carcinoma Progression. Int J Biol Sci 2022;18:3470-83. [Crossref] [PubMed]
- Yang Z, Yan C, Ma J, et al. Lactylome analysis suggests lactylation-dependent mechanisms of metabolic adaptation in hepatocellular carcinoma. Nat Metab 2023;5:61-79. [Crossref] [PubMed]
- Xiong J, He J, Zhu J, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell 2022;82:1660-1677.e10. [Crossref] [PubMed]


