Markers of resistance to anti-EGFR therapy in colorectal cancer
Introduction
Colorectal cancer (CRC) is the fourth most frequently diagnosed cancer and the second leading cause of cancer death in the United States accounting for 143,460 new cases and 51,690 deaths (1). At initial diagnosis, 40-50% of patients with CRC have metastatic disease highlighting the importance of effective systemic therapy (2). During the past 10 years, chemotherapeutic agents including oxaliplatin, irinotecan, cetuximab (CTX), panitumumab (PAM), bevacizumab, aflibercept and regorafenib (3-6), have been approved as an addition to the traditional fluorouracil (5-FU) treatment, doubling the median overall survival (OS) from approximately 12 to 22 months. The introduction of these agents has significantly increased the cost of care for CRC. In addition, these agents have potential for serious side effects. Both these considerations have raised the question of Biomarker development based on the mechanism of resistance as a method to select patients who would benefit from a specific therapeutic approach. The identification of Kirsten rat sarcoma-2 virus oncogene (KRAS) mutation as a mechanism of resistance to cetuximab (Erbitux®, Imclone, NY, USA) (7-9) and panitumumab (Vectibex®, ABX-EGF, Amgen, Thousand Oaks, CA, USA) (10-13), ushered the era of personalized medicine in CRC. Approximately, 40% of patients with CRC have KRAS mutations and are resistant to EGFR inhibitors. The presence of KRAS WT does not guarantee benefit from EGFR inhibitors; therefore other pathways of resistance and potential predictive biomarkers are greatly needed to identify the non-responders as well as those who will develop resistance after initial response in KRAS WT metastatic colorectal cancer (mCRC) patients.
Epidermal growth factor receptor (EGFR)
EGFR has been identified in many human epithelial cancers, including head and neck squamous-cell carcinoma, CRC, breast, pancreatic, non-small cell lung and brain cancer. EGFR is a glycoprotein of 170 kDa, encoded by a gene located on chromosome 7p12. The EGFR is a member of the human epidermal tyrosine kinase receptor (Her) family, which consists of EGFR (erbB1/Her1), Her2/neu (erbB2), Her3 (erbB3) and Her4 (erbB4). EGFR has an extracellular ligand-binding domain (domains I, II, III, IV), a single membrane-spanning region, a juxtamembrane nuclear localization signal and a cytoplasmic tyrosine kinase domain. Activation of the EGFR by ligands such as EGF, TGFα, amphiregulin, heparin-binding EGF, betacellulin and epiregulin in receptor hetero or homodimerization and activation of the tyrosine kinase domain. Phosphorylated cytoplasmic tails serve as docking sites for numerous proteins that contain src homology and phosphotyrosine-binding domains. EGFR activation stimulates complex intracellular signaling pathways that are tightly regulated by the presence and identity of the ligand, heterodimer composition, and the availability of phosphotyrosine-binding proteins. The two primary signaling pathways activated by EGFR are the RAS-RAF-MAPK and PI3K-PTEN/PTEN/AKT pathways (Figure 1). When activated, the PI3K/AKT pathway leads to protein synthesis, cell growth, survival, and mobility. The RAS/RAF/MAPK pathway leads to cell cycle progression and proliferation (14,15).
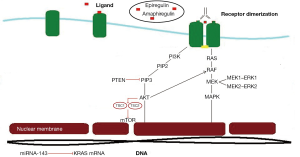
KRAS
The human homolog of the KRAS oncogene, encodes a small GTP binding protein that acts as a self-inactivating signal transducer by cycling from GDP- to GTP-bound states in response to stimulation of a cell surface receptor, including EGFR. KRAS can harbor oncogenic mutations that yield a constitutively active protein. Given that KRAS has a pivotal role in the transduction of EGFR signaling, evaluation of the impact of KRAS mutations as a mechanism of resistance to EGFR inhibition was a rational approach. Activating KRAS mutations in codon 12 are detected in approximately 35% to 45% of CRC (in the primary and metastatic site but not in lymph nodes). Several retrospective trials have demonstrated resistance to anti-EGFR targeted agents in patients whose tumors harbor the KRAS mutation (6,16). Summary of these trials are presented in Table 1. The role of KRAS mutation in resistance to EGFR inhibitors is best demonstrated in two pivotal trials that compared single agent EGFR inhibitor to best supportive care. In the first trial (NCIC trial), 572 patients with chemo-refractory disease were randomized to either CTX or best supportive care (BSC) as third line treatment. Cetuximab treatment was associated with a significant improvement in OS (HR, 0.77; 95% CI, 0.64 to 0.92; P=0.005) and in PFS (HR, 0.68; 95% CI, 0.57 to 0.80; P<0.001) (5). In a subgroup analysis, 394 of 572 patients were analyzed. Patients with KRAS WT demonstrated a statistically significant improvement in OS (median, 9.5 vs. 4.8 months; HR, 0.55; 95% CI, 0.41 to 0.74; P<0.001) and PFS (median, 3.7 vs. 1.9 months; HR, 0.40; 95% CI, 0.30 to 0.54; P<0.001). Patients with KRAS mutated tumors did not demonstrate any benefit in OS or PFS in CTX as compared to best supportive care (HR, 0.98; P=0.89) PFS (HR, 0.99; P=0.96) (27). Similar results were observed in a randomized trial that compared PAM to BSC in patients with chemo refractory mCRC. Patients with tumors that were KRAS WT had a significant improvement in PFS with a median of 8 weeks in the PAM compared to 7.3 weeks in the BSC group. (HR, 0.54; 95% CI, 0.44 to 0.66, P<0.0001). The objective response rates (RR) favored PAM over BSC, RR were 10% for PAM and 0% for BSC (P<0.0001). Patients with KRAS mutated tumors did not demonstrate any benefit on OS for PAM over BSC (HR, 1.00; 95% CI, 0.82 to 1.22) (26).
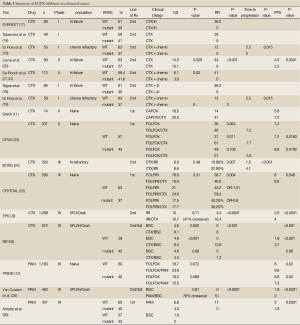
Full Table
In the CRYSTAL trial (Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer) by Van Custem et al., 1,198 previously untreated patients with advanced stage CRC were randomized to receive CTX plus FOLFIRI or FOLFIRI alone. It was found that there is a significant PFS advantage to the study combination (FOLFIRI/CTX) over FOLFIRI alone (HR, 0.85; 95% CI, 0.72 to 0.99; P=0.048). There was no significant difference in the OS (HR, 0.93; 95% CI, 0.81 to 1.07; P=0.31). In a subgroup analysis, patients whose tumors had KRAS mutation (37%), did not have any improvement in PFS (HR, 1.07; P=0.75) or OS (HR, 1.03) when CTX was added to FOLFIRI. Patients with KRAS WT tumors did demonstrate a statistically significant improvement in PFS with a median of 9.9 months compared to 8.4 months in the FOLFIRI alone group (HR, 0.68, 95% CI, 0.50 to 0.94; P=0.02), and OS (median of 23.5 vs. 20.0 months, HR, 0.84, P=0.0093) in favor of adding CTX to FOLFIRI. The RR was 57.3% vs. 39.7% (P=0.001) when compared to FOLFIRI alone. The results of this pivotal trial lead to the approval of CTX in the frontline setting in combination with FOLFIRI in patients with KRAS WT mCRC (6,25).
Activating mutations in codon 13 of the KRAS gene occur in about 6% of CRC. The role of codon 13 mutations in development of resistance to EGFR treatment is still controversial. An in vitro study showed that KRAS codon 13 mutations exhibit weaker transforming activity than codon 12 mutations in colon with low resistance to apoptosis and growth ability (28). DeRoock et al., studied the association between p.G13D mutation to response and survival in patients with chemotherapy-refractory CRC treated with CTX. The p.G13D-mutated tumors had longer OS of 7.6 months compared to 5.7 (P=0.005) and longer PFS (4.0 vs. 1.9 months; P=0.004). Although these results indicate that patients with p.G13D-mutated tumors respond to CTX, the results had a lower RR than patients with KRAS WT tumors. From the same study, in vitro and mouse model analysis showed that p.G12V mutated CRC cells were insensitive and p.G13D-mutated cells were sensitive, as were KRAS WT cells, to CTX (21). Peeters et al. evaluated the impact of KRAS codon 13 mutation status from three trials that evaluated PAM in advanced stage CRC. The results demonstrated that patient with tumors that harbor the KRAS codon 13 mutation do not benefit from PAM. Possible interpretations for the difference in effect of KRAS codon 13 mutation on sensitivity to EGFR inhibitors may include a difference between PAM and CTX or more likely a difference in the interaction of the codon 13 mutation with the chemotherapy backbone. At this point in the absence of prospective trials and given the contradictory results of the two retrospective studies, the role of KRAS codon 13 mutation in resistance to EGFR inhibition is still controversial (29).
Mechanisms of resistance beyond KRAS
Approximately half of patients with KRAS WT tumors do not respond to anti-EGFR treatment, raising the question of factors beyond KRAS mutational status that affect resistance. The potential factors include increased EGFR ligand expression, decreased EGFR expression, or activation of alternate signaling pathways.
Level of expression of EGFR, epiregulin and amphiregulin
Baker et al. analyzed biopsies from primary sites (validating the data from previous report of the metastatic site biopsy of the same group) for KRAS and EGFR ligand gene expression level. KRAS mutations were found in 43% of patients. In the KRAS WT setting, sensitivity to EGFR inhibition was proportional to the expression of EGFR ligands, epiregulin and amphiregulin. High ligand expression identified a subgroup of KRAS WT patients who had a high probability of responding to anti- EGFR compared to KRAS WT patients with low ligand expression who behaved like KRAS mutant CRC patients. In addition patients with high levels of the EGFR ligands were more likely to have disease control with CTX and significantly longer PFS than patients with low expression for both epiregulin (P=0.0002) and amphiregulin (P=0.0001) (30). There was no evidence of a relationship between epiregulin and amphiregulin gene expression and PFS and OS in patients with KRAS mutant tumors (31). In patients with high levels of mRNA for the EGFR ligands epiregulin and amphiregulin, CTX treatment tends to have a more potent antitumor activity. Therefore, the low expression of ligand may be a mechanism of resistance to EGFR inhibitors as it indicates that the EGFR system may not be the main contributor of tumor growth or progression.
EGFR expression
In the initial development of EGFR inhibitors, patients were selected to enroll on trials only if the tumors were positive for EGFR expression using immune-histochemistry. This was based on the concept that lack of EGFR expression, results in resistance to EFGR inhibitors. In the study by Chung et al., 4 of 16 patients with no EGFR expression, demonstrated significant responses to CTX based therapy (32). Therefore, EGFR expression using immune-histochemistry does not seem to impact resistance to EGFR inhibitors.
The role of EGFR expression in resistance to EGFR inhibitors was also evaluated using molecular based assays. In a study by Moroni et al., 31 patients with mCRC who had either a response or stable (30%) or progressive disease (70%) after CTX or PAM treatment were screened for EGFR gene copy number. Eight of nine patients with objective responses had an increased EGFR copy number. On the other hand, 1 of 21 non-responders had an increase in EGFR copy number (P<0.0001) (33). The same group assessed the role of EGFR copy number as a predictor of clinical outcome in patients treated with PAM. A mean EGFR gene copy number of less than 2.5/nucleus or less than 40% of tumor cells displaying chromosome 7 polysomy within the tumor, predicted a shorter PFS (P=0.039) and OS (P=0.015) (34). Lenz et al. also evaluated the effect of EGFR gene copy number on response to CTX using polymerase chain reaction (PCR) instead of the previously reported fluorescence in situ hybridization (FISH). Lack of association of increased gene copy number with objective responses and PFS but a positive correlation with OS was found (35). Retrospectively analyzed EGFR copy number by FISH from 85 samples of chemo-refractory mCRC patients treated with CTX, identified a positive EGFR FISH score that best associates with RR and longer time to disease progression when compared to EGFR FISH negative at a mean of 2.92 EGFR gene copy number (36,37). In the study conducted by Lievre et al., an increased EGFR gene copy number assessed by chromogenic in situ hybridization (CGH) was significantly associated with an objective tumor response to CTX. However, the low number of EGFR-positive patients precluded any firm conclusion (38). The largest investigation conducted in this regard, detected increased EGFR gene copy number at a frequency of 6% and found no association with disease control rate (33). A recent meta-analysis suggests that increased EFGR gene copy number is associated with improved survival from anti-EGFR treatment for mCRC patients (39). Overall, current data regarding the role of EGFR gene copy number as a mechanism of resistance to EGFR inhibition is inconsistent due controversial technique, uncertain level score cutoff, and lack of standardization. With the several methods used (FISH, qPCR, or CGH), it will be difficult to compare these studies.
BRAF
The serine-threonine kinase BRAF is the principal effector of KRAS. BRAF mutation is downstream to KRAS and is found in less than 10% of CRC. OS differs by somatic mutation status regardless of treatment received: BRAF mutant, 8.8 months; KRAS mutant, 14.4 months and KRAS WT, 20.1 months (40). BRAF V600E mutation indicated poor prognosis in patients with KRAS WT disease in FOLFIRI alone and FOLFIRI/CTX groups; those with BRAF mutations had worse outcomes. BRAF V600E mutations were detected in 6% of tumor samples. In nearly all cases, these mutations were identified in KRAS WT tumors and the impact of BRAF mutation in relation to efficacy of anti-EGFR was examined in the CRYSTAL trial population. The presence of BRAF mutation was a poor predictor of response and survival. Whether this biomarker is a negative predictor in relation to CTX is difficult to determine since this trial had a relatively small number of patients with BRAF mutations (6). In other trials, tumor with BRAF mutation was a negative prognostic marker for OS in patients with mCRC (41,42). In the NORDIC VII population, patients with mutated BRAF had low RR and markedly shorter PFS and OS compared to WT mutations (43). In a retrospectively analyzed study for endpoints of RR, time to progression, OS, and the mutational status of KRAS and BRAF, 113 tumors from CTX or PAM-treated mCRC patients were analyzed. The BRAF V600E mutation was detected in 14% of patients who had KRAS WT disease. None of the BRAF-mutated patients responded to anti-EGFR treatment and had significantly shorter PFS and OS compared to BRAF WT. The role of BRAF mutations in patients treated with EGFR-targeted drugs is similar to that of mutated KRAS (44). Furthermore, 50% of BRAF mutations are more frequently detected in microsatellite instability (MSI-high) CRC compared with microsatellite-stable 12% (45-47). Even with BRAF inhibition by vemurafenib, limited response has been defined. It is proposed that with this inhibition, more activation of the EGFR will result unlike melanoma cells which express low levels of EGFR on the cell surface (48-52). A cell-based analysis of a trial adding sorafenib to an anti-EGFR agent showed that even BRAF-mutated CRC cells can potentially respond to EGFR-targeted therapy if the BRAF inhibitor, sorafenib, is administered concomitantly with CTX or PAM even when either drug alone has limited activity. These data indicate that in BRAF-mutated tumors, the therapeutic effect of CTX or PAM could be restored by an approach aimed at blocking the EGFR pathway at multiple locations. In addition to sorafenib, other compounds targeting either BRAF (PLX4032) or its downstream effectors (ARRY-162, AZD6244, and PD0325901) are in clinical development and could be exploited in combination with EGFR-targeted moAb therapy (53,54). Despite KRAS and BRAF WT status, there have still been a significant percentage of non-responders (41%) to anti-EGFR therapy questioning further pathways that are important in defining resistance to these treatments (44).
PIK3K gene
In a study looking at chemo-refractory CRC patients treated with CTX and chemotherapy, for those with PIK3CA, exon 20 KRAS mutations had a worse outcome when compared to KRAS WT with a lower response rate and decreased median survival. PIK3CA mutations in exon 9 had no effect on survival and prognosis (40). Similar findings were seen in a review of the association between PIK3CA mutations and clinical outcomes of mCRC patients who were treated with anti-EGFR monoclonal antibodies (moAb); these results also suggest PIK3CA exon 20 may be a potential biomarker for resistance to anti-EGFR moAbs in KRAS WT mCRC (55). PIK3CA mutations have been associated with resistance to the anti-EGFR therapy since they can coexist with KRAS mutations; however it has been difficult to establish a definitive one-on-one relationship. Hot-spot mutations in PIK3CA mutations, specifically helical and kinase domain mutations, may operate by different but synergistic mechanisms independent of KRAS (56). However the role of PIK3CA mutation in EGFR resistance in mCRC patients remains controversial.
A study of PIK3CA in a group of 200 chemo-refractory mCRC patients who were treated with CTX in KRAS WT patients found no difference in CTX response in relation to PIK3CA status (57). PIK3CA mutations were detected in 16.4%. Only PIK3CA mutations occasionally coexisted with other gene mutations. In univariate analysis, prognostic significance for survival was seen for BRAF mutations codon 12-only KRAS mutations, high amphiregulin mRNA expression only in KRAS WT CRC, and high epiregulin mRNA expression regardless of KRAS mutation status. Favorable predictive factors were: high amphiregulin mRNA in KRAS WT tumors, high epiregulin mRNA, and low Ephrin A2 receptor mRNA. CTX-treated patients with amphiregulin-low KRAS WT CRC fared very poorly, with survival similar to KRAS mutant disease. Patients with KRAS codon 13 or other non-codon 12 mutations had a median survival similar to that of patients with KRAS WT; this is in contrast to patients with KRAS codon 12 mutations who did worse than all others (58). In terms of targeting treatment approaches, KRAS mutations show evidence of resistance to P13K pathway inhibitors (59). Specifically the presence of the mutant KRAS predicted resistance in the presence of the P13K inhibitor, PX-866 (60). This may limit the utility of single-agent P13K pathway inhibitors which have KRAS and PIK3CA mutations seen in colon cancers (61).
PTEN
Enhanced P13K signaling is often due to the activation of genes involved in the P13K pathway such as PIK3CA and AKT1, or loss of phosphatase and tensin homolog (PTEN) (62-64). Mutations in PTEN were seen in approximately 18% of patients with CRC tumors who had MSI suggesting that defective mismatch repair of PTEN may be a possible target for future therapies (65,66). Additional data suggests that PTEN promoter hypermethylation occurred frequently with high versus low MSI (19.1% vs. 2.2%; P=0.002) (67). A combined analysis of KRAS, BRAF, and PTEN showed increased RR in up to 45% for chemo-refractory patients receiving CTX from 39% with KRAS, PTEN and BRAF WT tumors where PTEN mutations were all resistant to CTX, unlike KRAS mutation where 12.5% in this study, responded to CTX (68).
MAPK
The intersection of KRAS-MAPK-PI3KCA pathway has direct implications for tumorigenesis. The rate of KRAS mutation was determined by sequencing exon 2, which has the most commonly mutated codons- codon 12 and 13 (69). Genetic variation in the MAPK signaling pathway affects colorectal cancer and may be affected by environmental and lifestyle factors including use of aspirin/NSAIDs, cigarette smoking, estrogen exposure and body mass index (70). Combination of P13K and MAPK pathway inhibition by treatment with a dual PI3K/mTOR inhibitor (NVP-BEZ235) and a MEK inhibitor (ARRY-142886) led to significant tumor regression in a KRAS lung cancer model (59).
MEK
Another downstream to KRAS target, is MEK. MEK activates extracellular signal-regulated kinases (ERK-1 and ERK-2) which are responsible for phosphorylation of factors that control cell cycle activation mainly at the G to S cell cycle progression. Resistance to EGFR-targeted therapy could also be mediated through alternate means of extracellular signal-regulated kinase 1/2 (ERK1/2) activation that bypasses EGFR either via alternative receptors at the plasma membrane or constitutively active downstream components. By generating CTX-resistant cell lines, Yonesaka et al. first identified multiple clones that exhibited less effective suppression of ERK1/2 phosphorylation in the presence of CTX. Further analysis of these clones revealed amplification of ERBB2 with corresponding increases in total and phospho-ERBB2 levels. Subsequent depletion of ERBB2 in the resistant clones restored sensitivity to CTX, confirming the importance of ERBB2 in the resistant phenotype. ERBB2 amplification is the proposed mechanism of CTX-resistant clones where acquired resistance was mediated by increased levels of heregulin, a ligand that binds ERBB3 and ERBB4. This leads to activation of downstream pathway targets and the role of this ligand is yet to be defined (71). In a recent molecular analysis, molecular changes to KRAS resulted in acquired resistance to anti-EFGR treatment. Mutant KRAS alleles treated with CTX were detectable ten months prior to radiographic evidence of disease progression. When combined with an EGFR inhibitor and MEK inhibitor early on, evidence suggests delay or reversal of drug resistance (72).
IGF1
The type 1 insulin-like growth factor receptor (IGF-1R) is a member of a family of trans-membrane tyrosine kinases that includes the insulin receptor and the insulin receptor-related receptor. The IGF-1R signaling pathway is important in different types of cancers and includes transduction of the IGF signal by the MAPK and PI3K/Akt. Preclinical data shows that combination treatment of IGF-1R and EGFR kinase inhibitors results in synergy of growth inhibition in CRC cell lines (73). Evidence suggests cross-talk between IGF-1R and EGFR, which might be crucial for the mitogenic and transforming activity of EGFR. More specifically, the IGF-1 downstream signaling cascade is thought to induce EGFR independent PIK3CA and AKT activity, which might be another explanation for the lack of efficacy of anti-EGFR monoclonal antibodies in KRAS WT CRC (74). This is supported by Bohula et al. in their experiments which proved that IGF-1 and IGF-2 are ubiquitously produced protein hormones that interact with the IGF-1 receptor (IGF1R) to regulate growth, differentiation, and survival. The IGF1R activates both RAS/ERK- and PI3K/AKT-related signal transduction pathways, which act to promote proliferation and prevent apoptosis (75). A phase II study with the anti-IGF-1R monoclonal antibody IMC-A12, either alone or in combination with CTX, was performed in patients with CTX or PAM-refractory mCRC. No antitumor activity was seen in the 23 patients treated with IMC-A12 monotherapy and of the 21 patients treated with the combination of IMC-A12 and CTX, 1 patient with KRAS WT achieved a partial response, with disease control lasting 6.5 months. No additional antitumor activity was observed in patients with the combination treatment (76). Concomitant blockade of IGF-1R and MEK has been shown to effectively prevent the occurrence of the EGFR-IGF1R cross-talk in BRAF mutated CRC preclinical models (77).
Conclusions
Despite the rapid advancement in EGFR targeted therapy, much remains to be studied to understand the mechanism of resistance in CRC. Clearly, KRAS codon 12 mutation is a leading cause of resistance to EGFR inhibitors. In the KRAS WT group several contributing factors appear to influence resistance and these include ligand expression, activation of the PI3K or IGFR-1 pathways. The role of RAS codon 13 mutations and BRAF mutations as a mechanism of resistance to EGFR inhibitors is an area that requires further research. Identification of mechanism of resistance to EGFR inhibitors will improve our ability to select patients for personalized medicine approach as well as develop new combinations of therapies that can overcome resistance to current available treatments.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012;62:10-29. [PubMed]
- Centers for Disease Control and Prevention: United States Cancer Statistics: US Cancer Statistics Working Group. Available online: http://www.cdc.gov/uscs, 2013.
- André T, Raymond E, de Gramont A. Regorafenib for metastatic colorectal cancer. Lancet 2013;381:1536-7. [PubMed]
- FDA approves aflibercept (Zaltrap) for metastatic colorectal cancer. Oncology (Williston Park) 2012;26:842, 873.
- Jonker DJ, O’Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040-8. [PubMed]
- Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011-9. [PubMed]
- Aboud-Pirak E, Hurwitz E, Pirak ME, et al. Efficacy of antibodies to epidermal growth factor receptor against KB carcinoma in vitro and in nude mice. J Natl Cancer Inst 1988;80:1605-11. [PubMed]
- Pollack VA, Savage DM, Baker DA, et al. Inhibition of epidermal growth factor receptor-associated tyrosine phosphorylation in human carcinomas with CP-358,774: dynamics of receptor inhibition in situ and antitumor effects in athymic mice. J Pharmacol Exp Ther 1999;291:739-48. [PubMed]
- Sobrero AF, Maurel J, Fehrenbacher L, et al. EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:2311-9. [PubMed]
- Yang XD, Jia XC, Corvalan JR, et al. Development of ABX-EGF, a fully human anti-EGF receptor monoclonal antibody, for cancer therapy. Crit Rev Oncol Hematol 2001;38:17-23. [PubMed]
- Borner M, Koeberle D, Von Moos R, et al. Adding cetuximab to capecitabine plus oxaliplatin (XELOX) in first-line treatment of metastatic colorectal cancer: a randomized phase II trial of the Swiss Group for Clinical Cancer Research SAKK. Ann Oncol 2008;19:1288-92. [PubMed]
- Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 2010;28:4697-705. [PubMed]
- Information on the conditional approval of panitumumab (Vectibix) by the EMEA. Available online: www.emea.europa.eu/pdfs/human/press/pr/32370307en.pdf 2011. (Accessed on April 20).
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127-37. [PubMed]
- Oda K, Matsuoka Y, Funahashi A, et al. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 2005;1:2005.0010.
- Han CB, Li F, Ma JT, et al. Concordant KRAS mutations in primary and metastatic colorectal cancer tissue specimens: a meta-analysis and systematic review. Cancer Invest 2012;30:741-7. [PubMed]
- Tejpar S, Humblet M, Vermorken Y, et al. Relationship of efficacy with KRAS status (wild type versus mutant) in patients with irinotecan-refractory metastatic colorectal cancer (mCRC), treated with irinotecan (q2w) and escalating doses of cetuximab (q1w): The EVEREST experience (preliminary data). J Clin Oncol 2008;26:4001. [PubMed]
- Tabernero J, Cervantes A, Ciardiello F. Correlation of efficacy to KRAS status (wt vs. mut) in patients (pts) with metastatic colorectal cancer (mCRC), treated with weekly (q1w) and q2w schedules of cetuximab combined with FOLFIRI. ASCO GI Cancers Symposium (January 25-27, 2008, Orlando, FL). 2008:abstr 435.
- Di Fiore F, Blanchard F, Charbonnier F, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer 2007;96:1166-9. [PubMed]
- Lièvre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008;26:374-9. [PubMed]
- De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 2010;304:1812-20. [PubMed]
- De Roock W, Piessevaux H, De Schutter J, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol 2008;19:508-15. [PubMed]
- Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 2011;22:1535-46. [PubMed]
- Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337-45. [PubMed]
- Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009;360:1408-17. [PubMed]
- Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 2007;25:1658-64. [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [PubMed]
- Guerrero S, Casanova I, Farré L, et al. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res 2000;60:6750-6. [PubMed]
- Peeters M, Douillard JY, Van Cutsem E, et al. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol 2013;31:759-65. [PubMed]
- Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007;25:3230-7. [PubMed]
- Baker JB, Dutta D, Watson D, et al. Tumour gene expression predicts response to cetuximab in patients with KRAS wild-type metastatic colorectal cancer. Br J Cancer 2011;104:488-95. [PubMed]
- Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005;23:1803-10. [PubMed]
- Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol 2005;6:279-86. [PubMed]
- Sartore-Bianchi A, Moroni M, Veronese S, et al. Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab. J Clin Oncol 2007;25:3238-45. [PubMed]
- Lenz HJ, Van Cutsem E, Khambata-Ford S, et al. Multicenter phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irinotecan, oxaliplatin, and fluoropyrimidines. J Clin Oncol 2006;24:4914-21. [PubMed]
- Cappuzzo F, Finocchiaro G, Rossi E, et al. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol 2008;19:717-23. [PubMed]
- Personeni N, Fieuws S, Piessevaux H, et al. Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clin Cancer Res 2008;14:5869-76. [PubMed]
- Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66:3992-5. [PubMed]
- Jiang Z, Li C, Li F, et al. EGFR gene copy number as a prognostic marker in colorectal cancer patients treated with cetuximab or panitumumab: a systematic review and meta analysis. PLoS One 2013;8:e56205. [PubMed]
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [PubMed]
- Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 2009;27:5931-7. [PubMed]
- Tol J, Dijkstra JR, Klomp M, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer 2010;46:1997-2009. [PubMed]
- Tveit KM, Guren T, Glimelius B, et al. Phase III trial of cetuximab with continuous or intermittent fluorouracil, leucovorin, and oxaliplatin (Nordic FLOX) versus FLOX alone in first-line treatment of metastatic colorectal cancer: the NORDIC-VII study. J Clin Oncol 2012;30:1755-62. [PubMed]
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705-12. [PubMed]
- Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res 2005;65:6063-9. [PubMed]
- Deng G, Bell I, Crawley S, et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res 2004;10:191-5. [PubMed]
- Wang L, Cunningham JM, Winters JL, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res 2003;63:5209-12. [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [PubMed]
- Ji H, Wang Z, Perera SA, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res 2007;67:4933-9. [PubMed]
- Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 2009;101:715-21. [PubMed]
- Vaughn CP, Zobell SD, Furtado LV, et al. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer 2011;50:307-12. [PubMed]
- Souglakos J, Philips J, Wang R, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer 2009;101:465-72. [PubMed]
- Solit D, Rosen N. Oncogenic RAF: a brief history of time. Pigment Cell Melanoma Res 2010;23:760-2. [PubMed]
- Migliardi G, Sassi F, Torti D, et al. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin Cancer Res 2012;18:2515-25. [PubMed]
- Mao C, Yang ZY, Hu XF, et al. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol 2012;23:1518-25. [PubMed]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 2008;105:2652-7. [PubMed]
- Prenen H, De Schutter J, Jacobs B, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res 2009;15:3184-8. [PubMed]
- Pentheroudakis G, Kotoula V, De Roock W, et al. Biomarkers of benefit from cetuximab-based therapy in metastatic colorectal cancer: interaction of EGFR ligand expression with RAS/RAF, PIK3CA genotypes. BMC Cancer 2013;13:49. [PubMed]
- Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008;14:1351-6. [PubMed]
- Ihle NT, Lemos R Jr, Wipf P, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res 2009;69:143-50. [PubMed]
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol 2010;28:1075-83. [PubMed]
- Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554. [PubMed]
- Ikenoue T, Kanai F, Hikiba Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res 2005;65:4562-7. [PubMed]
- Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997;275:1943-7. [PubMed]
- Guanti G, Resta N, Simone C, et al. Involvement of PTEN mutations in the genetic pathways of colorectal cancerogenesis. Hum Mol Genet 2000;9:283-7. [PubMed]
- Shin KH, Park YJ, Park JG. PTEN gene mutations in colorectal cancers displaying microsatellite instability. Cancer Lett 2001;174:189-94. [PubMed]
- Goel A, Arnold CN, Niedzwiecki D, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res 2004;64:3014-21. [PubMed]
- Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69:1851-7. [PubMed]
- Barault L, Veyrie N, Jooste V, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer 2008;122:2255-9. [PubMed]
- Slattery ML, Lundgreen A, Wolff RK. MAP kinase genes and colon and rectal cancer. Carcinogenesis 2012;33:2398-408. [PubMed]
- Yonesaka K, Zejnullahu K, Okamoto I, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med 2011;3:99ra86. [PubMed]
- Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532-6. [PubMed]
- Reinmuth N, Liu W, Fan F, et al. Blockade of insulin-like growth factor I receptor function inhibits growth and angiogenesis of colon cancer. Clin Cancer Res 2002;8:3259-69. [PubMed]
- Hu YP, Patil SB, Panasiewicz M, et al. Heterogeneity of receptor function in colon carcinoma cells determined by cross-talk between type I insulin-like growth factor receptor and epidermal growth factor receptor. Cancer Res 2008;68:8004-13. [PubMed]
- Bohula EA, Playford MP, Macaulay VM. Targeting the type 1 insulin-like growth factor receptor as anti-cancer treatment. Anticancer Drugs 2003;14:669-82. [PubMed]
- Reidy DL, Vakiani E, Fakih MG, et al. Randomized, phase II study of the insulin-like growth factor-1 receptor inhibitor IMC-A12, with or without cetuximab, in patients with cetuximab- or panitumumab-refractory metastatic colorectal cancer. J Clin Oncol 2010;28:4240-6. [PubMed]
- Buck E, Eyzaguirre A, Rosenfeld-Franklin M, et al. Feedback mechanisms promote cooperativity for small molecule inhibitors of epidermal and insulin-like growth factor receptors. Cancer Res 2008;68:8322-32. [PubMed]


