Personalized and precision medicine: integrating genomics into treatment decisions in gastrointestinal malignancies
Introduction
Cancer is a disease of the genome. In the past, limited insight into cancer biology hampered our understanding of the biology of disease and the development of targeted therapies. The advent of deep sequencing technologies has brought us to an unprecedented point in cancer history. The use of different sequencing platforms has facilitated identification of actionable genetic aberrations in different tumor types. This has fueled a number of clinical trials that target unique subsets of patients (baskets). Tumor heterogeneity is critical to understand response/resistance to therapy. Tumor heterogeneity is found not only among different patients (interpatient heterogeneity) but also among different sites of disease within the same patient (intrapatient heterogeneity) or even within a specimen from a unique site of disease (intratumoral heterogeneity). Still, the specific biological context in which genetic aberrations are found is relevant to the efficacy of a targeted therapy. This is illustrated by the failure of BRAF or PI3K inhibitors in BRAF or PIK3CA mutated metastatic colorectal cancer (CRC). In this context, the presence of epidermal growth factor receptor (EGFR) feedback loop or co-existing KRAS mutations may contribute to explain treatment failure. While basket studies are gaining momentum, failures remind us that shifting from a biology agnostic (histology-driven) approach to a histology-agnostic approach is unlikely to be a failure-free strategy for a number of tumor types.
Rationale for precision medicine
Although the terms personalized medicine and precision medicine are used interchangeably, differences exist between them. Personalized medicine is an older, broader term born from the Human Genome Project and refers to customization of treatment on the individual patient level (1). Precision medicine is a contemporary term that describes the utilization of molecular diagnostics to classify disease, and where possible, delivery of select treatment based on causal genetic variants (1). Current day molecular characterization of disease using next generation sequencing (NGS) enables a sensitive and specific diagnosis established by genotype. Correlating essential genotype with disease-modifying genes, environmental influences, and individual polymorphisms may help explain variations in phenotype (2).
The integration of genomics into clinical practice is transforming treatment paradigms. Identification of oncogenes and tumor suppressor genes can become the stimulus for rational design of novel, selective drugs that execute specific activity directed at underlying genetic aberrations. This is best exemplified by the success of imatinib in chronic myelogenous leukemia (3). Precision medicine has implications for furthering our understanding of cancer biology, explaining treatment-related failures and drug resistance, and guiding treatment planning. Multi-regional sequencing of tumors has revealed that malignancies exhibit genetic heterogeneity (4). Large-scale genomics projects have demonstrated that several molecular subtypes can exist within a single-tissue cancer type (5). Interrogation of these subtypes has revealed clinically important findings. One example is the identification of somatic mutations in exons 18–21 of the EGFR gene that are associated with sensitivity to tyrosine kinase inhibitor gefinitib (6). The existence of these somatic mutations sheds light on the observed gefitinib response in a subgroup of patients with non-small cell lung cancer (NSCLC) (7). In addition, this suggests there are inherent differences in EGFR-associated mutations in NSCLC compared to EGFR-associated deletions in gliomas where gefitinib has not demonstrated clinical activity (8). Similarly, BRAF mutations are predictive of response to BRAF inhibitors in patients with melanoma. However, single agent BRAF inhibitors have failed to show activity in BRAF mutated CRC, likely due to the release of a feedback loop through EGFR.
Tumor heterogeneity is likely to be underestimated and insufficiently captured with single tumor-biopsy analysis. Similarly, single biomarker-driven therapy may not be adequate, suggesting that targeting ubiquitous gene alterations may lead to better cancer control and patient outcomes (4).
In addition to transforming clinical practice and treatment landscapes, precision medicine is changing our perspective on the utility of clinical trials. Phase I trials were traditionally designed to evaluate drug dosing and establish safety. Now they also represent opportunities to exercise careful selection of patients who, based on genomic profiling, may benefit from biomarker-driven therapy. This finer method of identifying candidate patients for novel, genomically guided treatment offers a potentially more favorable benefit-to-risk profile (9).
The promises and challenges of matching genetic aberrations and targeted therapy
The integration of precision medicine into oncology has advanced research endeavors and clinical practice. Matching actionable aberrations with targeted therapy has led to survival improvement in different tumor types. Success stories offer hope that a molecular-based understanding of disease bridged with a molecular-driven approach to drug development can precisely match an individual’s genetic aberrations with a distinctive targeted therapy. Whether this genomically-guided approach to treatment broadly translates into favorable patient outcomes for patients with gastrointestinal (GI) cancer is still being studied.
A prospective, multi-center pilot study in the U.S. examined the effect of genomically-guided-based treatment on clinical outcomes (10). Among 86 of 106 patients in whom molecular profiling was attempted using immunohistochemistry, fluorescence in situ hybridization, or DNA microarray technology, a molecular target was determined in 84 cases (98%). Of these, 66 patients predominantly female with breast, colorectal, ovarian, or other types of rare cancers received genomically-guided treatment. With 18 patients (27%) receiving genomically-guided treatment and meeting the primary endpoint of progression free survival (PFS) ratio ≥1.3 (patients served as their own controls), it was concluded that a genomically-guided approach is feasible and promising. Study limitations included possible ascertainment bias from frequency of tumor evaluation influencing the PFS ratio, lack of randomization, and attrition bias.
In another study that reported on a single-institution’s experience with a personalized medicine program in the setting of phase I trials, the clinical outcomes of 175 patients with genomically-guided treatment were compared with those of 116 consecutive patients without genomically-guided treatment (11). The top three cancers were melanoma (73%), thyroid (56%), and CRC (51%). The top three molecular aberrations were RET (56%), TP53 (37%), and KRAS (18%). Among patients with one molecular aberration, genomically-guided treatment improved overall response rate (RR) (27% vs. 5%, P<0.0001), prolonged time-to-treatment failure (5.2 vs. 2.2 months, P<0.0001), and increased survival (13.4 vs. 9.0 months, P=0.017). This study showed benefit of therapy matched to molecular aberration. Limitations included the retrospective study design, absence of randomization, treatment response from a targeted agent combined with cytotoxic therapy, and most importantly, results were predominantly driven by patients with BRAF mutated melanoma (12).
More recently, a multi-center study in France randomized adult patients with metastatic solid tumors refractory to standard of care to either treatment with a genomically-guided targeted agent (GGT, n=99) or treatment by physician’s choice (control, n=96). Targeted agents included erlotinib, lapatinib plus trastuzumab, sorafenib, imatinib, dasatinib, vemurafenib, everolimus, abiraterone, letrozole, or tamoxifen. The primary endpoint was median PFS. After a median follow-up of 11.3 months in both cohorts, no significant difference in PFS was observed: 2.3 months in the GGT group vs. 2.0 months in the control group (P=0.41). No difference in grade 3–4 adverse events was seen between both groups (P=0.30) (13). Overall, this study demonstrated that genomically-guided treatment is not superior for benefit in PFS compared to physician-choice treatment. These results bring into question the validity, utility, and value of genomically-guided personalized medicine in an era where NGS is increasingly being utilized. It should be pointed out that these negative results may stem from suboptimal drug selection per the study’s treatment algorithm.
A first-of-its-kind precision medicine trial is the National Cancer Institute (NCI) MATCH study (14). This trial has national reach and is most unique in its basket trial approach to treatment: it is purely based on genetic aberration. Thus, a patient with a given genetic abnormality may be matched to a precision medicine therapy regardless of malignancy. Another prospective precision medicine clinical trial is the Targeted Agent and Profiling Utilization Registry Study (TAPUR) available at several cancer center networks throughout the U.S. (NCT02693535). Although both innovative NCI MATCH and TAPUR studies are investigating molecularly-targeted cancer drugs and evaluating resultant clinical outcomes, they differ by geographical access to therapy, range of targeted therapies offered, and endpoints. Nevertheless, both studies evince the growing number of promising targeted oncology agents and the urgency to find more efficient ways to study these agents. Conversely, the underlying molecular reasons for treatment response, particularly to chemotherapy, are being studied through the NCI Exceptional Responders Initiative (15). Exceptional responders are defined as those having a complete response to therapy, those who have a partial response lasting >6 months or those who respond to a therapy for which the RR is less than 10%. This study approach can potentially identify key molecular characteristics of tumors associated with exceptional response. An important limitation of this study is that a report with actionable information is not returned to the treating physician.
Molecular profiling
NGS technologies
NGS refers to a group of technologies that can massively parallel sequence millions of nucleotide templates, which is the evolution of the first-generation dideoxy “Sanger” sequencing. The huge impact of NGS technologies on genetics and clinical medicine in the past several years has been described in several reviews (16-18). The high throughput of NGS has not only the capacity to produce an enormous amount of sequence data at much lower cost per base pair, but also to comprehensively detect all classes of genomic alterations including single nucleotide variation (SNV), short insertion/deletion (indel), copy number variation (CNV) and gene fusion/genomic rearrangement structure variation (SV) by testing on one sample. The most common platforms for NGS include Illumina products (HiSeq/MiSeq/NextSeq/MiniSeq) and Life technologies products (Ion PGM/Proton, ThermoFisher). The major NGS platform from Illumina uses the sequencing-by-synthesis (SBS) method for sequencing (19,20). SBS is an optical technology with distinct fluorescently labeled nucleotides to visualize the synthesized strand. In contrast, Ion PGM/Proton is semiconductor-based belonging to non-optical technology (21). The Ion semiconductor-based technology also uses SBS, where sequencing is performed in microscopic wells interfaced with a semiconductor chip. Instead of detecting optical signals, the semiconductor chip measures the change in pH caused by the protons released from the 3’-OH group during formation of phosphodiester bonds. Despite the numerous research and clinical applications reported on the Ion platform, it has relatively higher false positive rates and lacks the capability for long repeat sequencing detection compared with Illumina platforms (Table 1) (22-25).
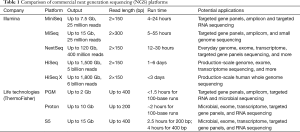
Full table
Clinical application of NGS
NGS shows a significant impact on diagnosis, management, and treatment of cancer. NGS costs less than Sanger sequencing for multi-gene panels and performs better on mutations with low mutant allele frequency (26-29). The Cancer Genome Altas (TCGA) projects, which took almost 10 years to finish using targeted sequencing, catapulted our understanding of the molecular basis of cancer and also provided many potential therapeutic targets. According to the sequencing results, many well-known actionable driver genes such as EGFR were demonstrated in multiple cancer types besides those previously characterized. Many cases were also reported to have actionable mutations that showed response to targeted therapies (30,31).
NGS testing can be performed on tumor tissue or blood
NGS testing can reveal somatic variation due to tumor heterogeneity. Therefore, not all tumor cells may carry a particular somatic genetic variation requiring higher sensitivity for the sequencing technologies and biologic samples with a high content of tumor (generally 20%). Thus, multi-gene panels with ultra-deep coverage may be more suitable in oncology than whole exome sequencing. Blood is a convenient sample source and usually referred to as a “liquid biopsy” for isolating circulating tumor DNA (ctDNA) shed into systemic circulation from primary and metastatic disease sites. Blood is a desirable sample source due to the ease of attainment, as many patients have disease that is not amenable or safe to biopsy. Studies have demonstrated a high consistency of results from ctDNA compared to tissue testing; however, continued correlative studies are necessary (32). Blood also allows for serial sampling to detect mutations present at diagnosis and those that are acquired during the disease course or treatment. Serial ctDNA sampling may allow for earlier detection of drug resistance and also aid in monitoring for disease response. It is unclear that liquid biopsies will have an impact in disease management other than for patients with lung cancer. For instance, a recent study showed that less than 44% (74/168) of patients who underwent liquid biopsies had potentially actionable aberrations (33). More importantly liquid biopsies only influenced the treatment of two patients with NSCLC and one patient with breast cancer for which EGFR and PIK3CA mutations had already been identified in tissue. In addition, there are a number of limitations that need to be considered. Genetic aberrations found in liquid biopsies in elderly patients may originate from undiagnosed myelodysplastic syndromes rather than the suspected tumor. This was recently shown in a patient with metastatic pancreatic cancer where a JAK2 mutation was found in liquid biopsy, not previously described on TCGA and not identified on tumor tissue (34). Furthermore, the role of liquid biopsies in monitoring response to therapy remains to be validated. In the absence of disease progression by Response Evaluation Criteria In Solid Tumors (RECIST) there is no evidence to support switching therapies based on increased allele copy number of a given mutation. Currently, there are few publications on multi-gene panel sequencing for ctDNA, yet there is potential direction for the clinical application of NGS technologies. More details about tissue and ctDNA detections are shown in Table 2 (35,36).

Full table
Integrating genomics into treatment decisions in GI malignancies
As we navigate the intricacies of the human genetic blueprint and its role in cancer pathogenesis, one of the goals is to find genomic alterations that may have therapeutic potential. Among the 12 anti-cancer medications approved in 2015, more than half were classified as precision therapies (37). Thus, molecular drivers of cancer will continue to be the foundation for future investigations, new therapies, and new indications for existing therapies. Here, we summarize the current literature on genomics in GI malignancies.
Cholangiocarcinoma (CCA)
CCA arises from bile duct epithelium. It is the second most common hepatic malignancy worldwide and the fifth leading cause of death by cancer in US (38,39). This is a heterogeneous group of diseases that includes gallbladder cancer, intrahepatic CCA (IHCCA), and extrahepatic CCA with diverse molecular profiles and clinical course. Currently, the role of molecular profiling in CCA is not established. Since up to two-thirds of patients with CCA may harbor genomic alterations associated with targeted therapies, a molecularly-driven approach is promising (40-42). In IHCCA, IDH1/IDH2 mutations are unique (20%) and fibroblast growth factor receptor (FGFR) translocations (10%) are common. HER2 amplifications are more prevalent in extrahepatic cholangiocarcinoma and gallbladder cancer compared to IHCC (11–16% vs. 3%) (43-45). The development of therapeutic agents are directed at several key pathways, including tyrosine kinases, mutant isocitrate dehydrogenase (IDH) enzymes, PI3K-AKT-mTOR pathway, and chromatin remodeling networks (46,47). Of growing interest is the IDH gene, which has been described as the first gatekeeper in carcinogenesis (48). The IDH mutation is prevalent in intrahepatic HCC and affects “oncometabolite” 2 hydroxyglutarate involved in regulating histone methylation and DNA modification (45,48,49). In vitro studies have shown that mutant IDH can suppress hepatocyte differentiation and promote development of IHCCA (50). Increased levels of TP53 and abnormal levels of DNA methylation are common features of mutations in IDH1 (codon 132) and IDH2 (codon 140 and 172) (48). Although it is unknown if mutant IDH is required for tumor maintenance, it may nevertheless be a potential therapeutic target for patients with this distinct genetic subtype (50). The prognostic role of IDH1/2 mutations is conflicting with some reports demonstrating no prognostic significance and others associating IDH with longer survival. These discrepant findings warrant further study and validation (45,46,49,51). Presently there are a few trials in solid tumors, predominantly in phase I development, examining agents for IDH mutations (Table 3). ClarIDHy is a placebo-controlled, randomized phase III study testing AG-120 in patients with IDH1 positive CCA that has progressed to gemcitabine or 5-fluoruracil. The primary endpoint is PFS (52).
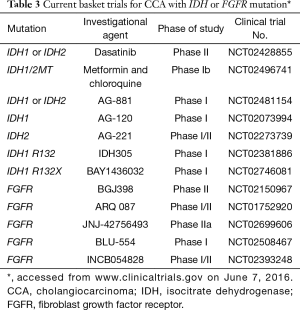
Full table
Different classes of IHCCA based on molecular profile have been proposed, suggesting that different treatment paradigms may be needed (53). Another promising targetable genomic alteration is the FGFR mutation present in 14% to 45% of IHCCAs (54). FGFR2 kinase fusion genes, FGFR2-AHCYL1 and FGFR2-BICC1 relevant to cancer transformation by activation of the MEK pathway, were detected using whole genome and transcriptome analyses (40,55-57). Overexpression of FGFR induces cell proliferation. Treatment of FGFR fusion-positive lines in bladder cancer cells in vitro with FGFR inhibitors PD173074 and multi-targeted TKI pazopanib demonstrated sensitivity and inhibition of cell proliferation (57). In a phase I study JNJ-42756493, another investigational pan-FGFR inhibitor, exhibited clinical activity in advanced solid tumors (58). Most recently in a phase II study, pan-FGFR inhibitor BGJ398 demonstrated an overall disease control rate of 82% in patients with advanced FGFR-altered CCA (FGFR2 fusions or other FGFR aberrations) who had progressed on cisplatin and gemcitabine chemotherapy. Noteworthy yet manageable side effects included hyperphosphatemia (an on-target class side effect), fatigue, constipation, cough, and nausea (59). Current investigations of IDH and FGFR mutations will hopefully translate into the development of effective targeted therapies (Table 3).
CRC
Cancer of the colon and rectum ranks in the top three newly diagnosed cancers and cancer-attributed deaths worldwide (60,61). Pivotal studies of anti-EGFR therapy (cetuximab, panitumumab) independently and unequivocally have demonstrated that patients with metastatic KRAS-mutated CRC do not benefit from these therapies (62,63). In addition, retrospective analysis have shown lack of benefit in tumors harboring any RAS family mutation. Current guidelines support expanded RAS testing (KRAS and NRAS exon 2, 3, and 4) in metastatic CRC (64). Thus, testing of RAS has important treatment implications. Patients with mutation in RAS genes should not be treated with EGFR antagonists (65). In addition, cetuximab and panitumumab have been shown to be suboptimal therapies in NRAS-mutated CRC (66). Given the negative predictive value of RAS mutations, the goal of genomic testing for RAS testing is to identify patients (wild-type) who will most likely have a favorable benefit-to-risk profile from treatment with cetuximab or panitumumab.
In the RAS/RAF/MEK/ERK signaling pathway, BRAF lies downstream from KRAS and requires phosphorylation to become activated. The role of BRAF as a negative predictor of response to anti-EGFR therapy has not been consistently evidenced across randomized clinical trials. Still, some studies indicate that patients with BRAF V600E-mutated metastatic CRC do not respond to anti-EGFR therapy, and none of the patients who responded to either of these treatments had BRAF mutations (67-69).
However, retrospective studies suggest clinical benefit with cetuximab in patients with BRAF V600E-mutated metastatic CRC, which is typically associated with poor prognosis (70). Thus, while BRAF is a strong prognostic marker, its role as a predictive marker remains unclear. Additionally, vemurafenib (BRAFi) failed to show activity as single agent in BRAF mutated metastatic colorectal cancer (mCRC) (71,72). Intriguingly, studies in vivo and in vitro have demonstrated that BRAF inhibition disrupts a negative feedback loop through EGFR, which subsequently promotes cellular proliferation. Based on this proposed mechanism, patients with BRAF V600E-mutated metastatic CRC may benefit from combination therapy consisting of both a BRAF inhibitor and an EGFR inhibitor (73). Currently, ongoing studies are examining dual blockade of BRAF with vemurafenib and EGFR with cetuximab plus irinotecan in patients with BRAF V600E mutated advanced CRC (NCT02164916) (Table 4) (74). Recent report from SWOG 1406 indicates that the addition of vemurafenib to irinotecan and cetuximab versus irinotecan and cetuximab produced greater median PFS (4.4 vs. 2.0 months), RR (16% vs. 4%), and disease control rate (67% vs. 22%, P<0.001) (75). Other mechanisms that may be contributing to the resistance phenomenon with anti-EGFR therapies include HER2 and mesenchymal epithelial transition (MET) aberrations. HER2 mutation and amplification is reported in 7% of CRC (76). Overexpression of HER2 has been implicated in resistance to anti-EGFR therapy via aberrant MEK-AKT pathway activation (77). Dual HER2 inhibition with trastuzumab and lapatinib in patients with HER2-amplified, KRAS wild-type metastatic CRC who are resistant to standard therapies, including EGFR-targeted agents, demonstrated that dual HER2 blockade can produce objective responses (78). These findings suggest a need for continued exploration in order to find viable treatment options for HER2-positive CRC (78). Amplification of the MET proto-oncogene (MET) is another potential mechanism that leads to acquired resistance in tumors that do not develop KRAS mutations during anti-EGFR therapy (79,80). In addition, patients with advanced CRC with PIK3CA mutations have been observed to have worse PFS, suggesting that PIK3CA mutations can independently hinder response to EGFR inhibitors (81). However, the independent predictive role of PIK3CA has not been consistently observed and the discrepancy may be related to the aberration present in exon 20 (82-84).
The canonical pathway in CRC is wingless-int (Wnt) signaling, particularly involving inactivating mutations of adenomatous polyposis coli (APC) or abnormal β-catenin-dependent gene expression (85). More than 80% of colonic adenomas and carcinomas have APC gene mutations, where loss of APC function leads to constitutive activation of Wnt signaling (86). Therefore, therapies targeted at Wnt represent a rational and potentially effective approach to treatment of CRC (77,86). Small molecules targeting the cancer stem cells of the Wnt signaling pathway at the extracellular, cytoplasmic, or nuclear levels may have potential as CRC therapies (87). In addition, there may be cross talk between the Wnt and EGFR pathways in tumorigenesis (85). Thus, simultaneous blockade of Wnt (WNT974), EGFR (cetuximab), and BRAF (LGX818) may be an effective combination treatment and is currently being studied (NCT02278133) (88). These studies reflect the complexity of the molecular pathogenesis of CRC. Genomic studies continue to be important not just for revealing potential new therapeutic targets but also to help explain lack of response and mechanisms of resistance to current targeted therapies in CRC.
Lastly, microsatellite instability (MSI-high) appears to confer immunogenicity due to presence of neoantigen-specific tumor infiltrating lymphocytes. Less than 5% of patients with mCRC are considered MSI-high. To counteract this inflammatory microenvironment, MSI-high mCRC tumors selectively upregulate the expression of immune checkpoint molecules, such as PD-1, PD-L1, CTLA-4 and LAG-3 (89). A phase II study evaluating immune checkpoint blockade with pembrolizumab in patients with advanced GI malignancies demonstrated that mismatch-repair status predicted clinical response (90). These results support a genetically-guided approach to immunotherapy in this unique subset. Currently, checkpoint inhibitors approved for different indications as well as novel checkpoint inhibitors (TIM-3, Lag-3, OX40, GITR, 4-1BB, CD40, CD70) are being studied in CRC (91). Recent work showed that MEK inhibition promotes infiltration of effector CD8+ T cells and synergize with a PD-L1 inhibitor in preclinical models (92). Indeed, a phase I study recently tested cobimetinib (MEK inhibitor) with atezolizumab (PD-L1 inhibitor) in MSI-low mCRC (93). The study showed a promising RR of 17% in this refractory population. These results warrant further evaluation and could open new treatment opportunities for patients with mCRC, the majority of whom have MSI-low tumors.

Full table
Gastroesophageal cancer (GEC)
In the U.S., GEC represents 1% of all new cancer cancers and 2.6% of all cancer deaths (94). The 5-year survival rate is 18%. Worldwide gastric cancer ranks among the top five cancers to affect males and females, and esophageal cancer is the 7th most common cancer in males (60). Presently, there are two targeted therapies approved in GEC, trastuzumab and ramucirumab. Trastuzumab is approved as first line treatment combined with cisplatin and capecitabine or 5-fluorouracil. In patients with HER2/neu-positive advanced GEC who were treated with trastuzumab and chemotherapy (cisplatin plus fluorouracil or capecitabine), there was an improved overall survival (OS) (median 13.8 vs. 11 months, P=0.0046) compared to patients treated with chemotherapy alone. Addition of trastuzumab was tolerable, and there was no difference in grade 3 or 4 or cardiac adverse events. The majority of patients in this study had gastric cancer (~80%), and post-hoc analysis showed survival benefit with tumors having IHC 3+ or IHC 2+ with FISH amplification compared to those with low expression of HER2 (95). HER2 amplifications vary by histology, and appear to be more prevalent in intestinal type than diffuse type (96,97). A prospective study in patients with advanced gastric cancer showed that the level of HER2 amplification indicated by a HER2/CEP17 ratio of 4.7 was more likely to predict response to therapy (96).
Four gastric cancer subtypes have been identified through molecular interrogation: Epstein-Barr virus (EBV)-positive type, MSI-high, chromosomally stable [low number of somatic copy number alterations (SCNAs), and chromosomally unstable (high number of SCNAs] (98). Genomic characterization of gastric carcinoma has revealed that TP53, PTEN, CTNNB1, ARID1A, KMT2C, and FAT4 genes are frequently mutated (98,99). ARID1A which is involved in chromatin remodeling was highly represented in 83% of gastric cancers with microsatellite instability and 73% of EBV-positive cases (100). Specific to esophageal adenocarcinoma, the top five significantly mutated genes have been reported to be TP53, CDKN2A, SMAD4, ARID1A, and PIK3CA (101). Comprehensive genomic profiling has revealed that there are more frequent KRAS and ERBB2 genomic alterations in esophageal adenocarcinoma vs. more PIK3CA, PTEN, and NOTCH1 alterations in esophageal squamous cell carcinomas (102). Understanding the molecular underpinnings has implications for treatment, including: PI3K inhibitors, programmed cell death targeted therapies, and JAK2 inhibitors for the EBV subtype; PI3K inhibitors, anti-HER3 therapy, and programmed cell death targeted therapies for tumors with microsatellite instability; anti-HER2 or anti-HER3 therapy, VEGF2-receptor antagonist, FGFR kinase inhibitor, and MET TKI for chromosomally unstable tumors (99). In addition, patients with metastatic gastric cancer with low levels of ATM exhibited survival benefit from the additive treatment effect of olaparib, a PARP inhibitor, combined with second-line paclitaxel (103).
In GEC a promising therapeutic target is the hepatocyte growth factor (HGF) ligand/MET factor receptor signaling axis involved in cell migration, invasion, and angiogenesis. MET expression has been seen in 24% of gastric carcinoma cells. High level MET expression (IHC 3+) is associated with lymph node metastases, more advanced stage of disease, and inferior survival (104). MET amplification is estimated to occur in 30% of gastric cell lines and similarly associated with poor prognosis (105-107). MET proto-oncogene has been shown to be an independent prognostic factor in gastric carcinoma, and is a potential molecular marker for targeted therapy (107,108). In vitro studies have demonstrated that MET kinase inhibitor PHA-665752 causes arrest in cell proliferation and apoptosis in gastric cancer cells with constitutively activated MET whereas gastric cancer cells without constitutively activated MET were unaffected (109). Currently, AMG102 (rilotumumab) and MetMAb and PRO143966 (onartuzumab) are under active investigation as MET-targeting agents (56,110-112). New results from a phase III study of AMG102 indicate that patients with advanced GEC treated with AMG102 adjunct to standard chemotherapy did not have improved OS compared to patients treated with chemotherapy alone. As a result, the manufacturer has ceased all phase III studies examining AMG102 in gastric cancer (113). In addition, MetMAb added to standard of care in patients with HER2-negative, MET-positive GEC did not confer survival benefit. Subgroup analysis indicates that non-Asian patients and patients without gastrectomy may benefit from MetMAb (114). Overall, it appears that MAB therapy does not work in gastric cancer, although there remains promising data with MET TKI therapy. For example, the observation that MET-independent HER kinase activation is a possible mechanism of MET TKI resistance in MET-amplified tumor cells that co-express EGFR and/or HER-3 is intriguing and suggests that combined pathway inhibition may be required (115). There remain limited treatment options in advanced GEC highlighting further investigation is needed in this area.
Hepatocellular carcinoma (HCC)
Liver cancer is on the rise, and projected to become the third-most common cancer by 2030 (116). HCC accounts for 90% of liver cancers (117). In the U.S. it is responsible for 2.2% of new cancer cases and 4.2% of cancer-related deaths. The 5-year survival is estimated to by 17.2% (118). HCC worldwide is among the top ten cancers for both incidence and cancer-related mortality (61). Presently, the only approved molecularly-based therapy in HCC (unresectable) is sorafenib, an oral small multikinase inhibitor of Raf, VEGFR-2 and -3, PDGFR β, c-KIT, fms-like tyrosine kinase (FLT)-3, and rearranged during transfection (RET). In the registry trial, patients with advanced HCC and Child-Pugh A treated with sorafenib 400 mg twice-daily had improved OS (median, 10.7 vs. 7.9 months, P>0.001) and time to progression (TTP) (median, 5.5 vs. 2.8 months, P<0.001) compared to patients receiving placebo (119,120). In 2011, sorafenib catalyzed almost 200 trials examining 56 different molecular-based therapies (121). Unfortunately, studies in both first-line and second-line settings evaluating other targeted agents or combination therapy with sorafenib have produced disappointing results (Table 5). Most recently, a phase III trial demonstrated survival benefit with second-line regorafenib after sorafenib failure when compared to placebo (median 10.6 vs. 7.8 months) (122).

Full table
The grim prognosis of advanced HCC reflects the aggressive nature of this cancer and the dearth of effective systemic treatments (123). Chemotherapy with fluorouracil, cisplatin, and/or gemcitabine offers modest survival benefit (124). NGS exploration of HCC has revealed genetic and epigenetic alterations, such as methylation, that affect p53/RB, Wnt/β-catenin, PI3K/PTEN/Akt/mTOR pathways (125,126). These pathways are involved in deregulation of oncogene and tumor suppressor genes and ultimately the development and progression of HCC. The most frequent mutations affect TERT promoter, TP53, CTNNB1. Less frequently mutated genes include AXIN1, ARID2, ARID1A, TSC1/TSC2, RPS6KA3, KEAP1, MLL2 (127). Currently, some of the targeted therapies for HCC being studied in open trials include small molecule MET inhibitors (tivantinib, INC280, cabozantinib, MSC2156119J), STAT inhibitor (OPB-111077), mTOR inhibitors (temsirolimus, MLN0128), and monoclonal antibodies (BIIB022, cetuximab, bevacizumab, pembrolizumab, ramucirumab, nivolumab, TRC105). These investigations will hopefully further address the high unmet need in HCC for therapeutic molecularly-based targeted therapies.
Pancreatic cancer
Pancreatic ductal adenocarcinoma (PDAC) is the fourth most common cause of death from cancer in the U.S. (38). In 2016, 53,070 new cases will be diagnosed and 41,780 patients will succumb to the disease. It is estimated that PDAC will be second only to NSCLC as the leading cause of cancer-related mortality in the USA by 2030 (116). Most PDAC patients present with advanced disease, and therefore they are not candidates for surgical resection. Palliative chemotherapy in these patients leads to modest improvements in survival (128). Erlotinib, an EGFR TKI, is the only targeted therapy approved by the FDA in this disease. However, the survival improvement when erlotinib was added to gemcitabine in a randomized phase III study was negligible (129,130). Phase III studies examining other molecularly-guided agents, axitinib, bevacizumab, cetuximab, ziv-aflibercept, added to gemcitabine-based chemotherapy have demonstrated absence of survival benefit (131-135).
PDAC is characterized by an average of 63 genetic alterations, mainly point mutations, involving 12 core signaling pathways. However, pathway components vary among individual tumors and can further present treatment challenges (136). Whole genome analysis recently identified four subsets of PDAC (137). Tumors in the subset known as “unstable” were associated with a BRCA signature as well as mutations in the DNA repair pathway including BRCA 1/2, PALB2 (partner and localizer of BRCA2) and ATM (ataxia telangiectasia mutated). This subset was more likely to respond to platinum based therapies. This subset may also benefit from PARP inhibitors. PARP are nuclear enzymes that regulate repair of DNA breaks through base-excision. PARP inhibitors are synthetically lethal in tumors with defective DNA repair (138). A phase II study of olaparib [poly(ADP-ribose) polymerase (PARP) inhibitor] in patients with solid tumors with germline BRCA1/2 mutations, showed that 5/23 (22%) patients with PDAC had partial response to olaparib (139). This has led to an actively recruiting phase III study comparing olaparib with placebo in patients with germline BRCA1/2 mutated metastatic PDAC and stable disease after first-line platinum therapy (NCT02184195) (140). Studies testing novel PARP inhibitors such as ABT-888 (veliparib), AGO14699 (rucaparib), and BMN-673 (talazoparib), are also currently underway (139,141-143).
A mutation of growing interest is tumor suppressor gene PALB2. PALB2 also known as FANCN, is most commonly known for its association with Fanconi anemia. In hereditary or sporadic pancreatic cancer (PC), PALB2 mutation appears sufficient to cause DNA replication and damage response defects (137,144-148). Several independent case studies have validated PALB2 as a susceptibility gene and actionable therapeutic target (144,149). Overall, two mutations of PALB2 have been identified as PC-specific: c.508_509delAG and c.3256C>T (150). PALB2 is a key player in maintenance of genomic integrity, and its full potential as a therapeutic target has yet to be uncovered.
In addition, a small subset of patients with advanced PDAC present microsatellite instability (<4%) (151). These tumors have deficient mismatch repair and subsequently increased mutation load. A small study recently reported two partial responses among four subjects with MSI-H PDAC. Remarkably, responses were durable (152). Basket studies with checkpoint inhibitors in MSI-H PDAC are planned. NGS has identified actionable aberrations at low prevalence (<5%) in a number of additional genes in this disease including among others: HER2, MET and PIK3CA (152-154). Drugs targeting these aberrations have already been approved for different indications. Basket studies targeting these subsets are warranted.
The study of familial pancreatic cancer has revealed additional opportunities for precision medicine in this disease. For instance, patients with Peutz-Jegher disease harbor a germline mutation in a tumor suppressor gene known as serine/threonine kinase 11 (SKT11). Loss of STK11 leads to constitutive activation of the mTOR pathway. Thirty-five percent of subjects with this disease will develop PDAC.
In an interesting case study, a patient with Peutz-Jeghers syndrome-induced PC treated with mTOR inhibitor everolimus achieved partial remission of acinar cell PC and concomitant clearance of large colon polyps (155). Unfortunately a study testing everolimus in patients with Peutz-Jeghers syndrome had to be closed due to poor accrual (NCT01178151). Finally, tissue acquisition remains to be a challenge to fulfill the promise of precision medicine in this disease as many of these patients are diagnosed with fine needle aspirations (FNA) rather than core biopsies. The IMPaCT Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) study recently showed that precision medicine can be implemented in real time in this disease with over 75% of patients having sequencing results available within 4 weeks (146). Future studies will expedite turnaround times and will contribute to elucidate if this strategy will improve outcomes.
Conclusions
GI malignancies are clinically and molecularly heterogeneous. Actionable aberrations are present at low prevalence across different tumor types. These aberrations define unique molecular subsets within each cancer type. Treating patients with drugs matching the unique genetic aberrations found in their disease has the potential to improve outcomes. However, this will need to be tested in clinical trials. Ultimately, the proof of concept will be to demonstrate that this precision medicine strategy improves survival. Several challenges will need to be overcome to achieve this. First and foremost, there is a limited availability of clinical trials to enroll patients once an actionable aberration has been identified. For instance, in the IMPaCT study only 1 out of 34 screened PDAC was able to proceed with matched therapy. Allocating resources to coordinate patient travel to a study center close to home is a strategy that has been implemented in the STARTRK-2 study (NCT02568267) (156). Second, appropriate validation of biomarkers of response as well as drug selection are needed. In this regards, we hypothesize that negative results of SHIVA study could be related to drug selection. Third, the use of liquid biopsies, while clearly representing an opportunity to address real-time tumor heterogeneity, needs further validation. In particular, we will need to define if prospective collection of samples such as plasma and urine, to detect early emergence of resistance to therapy, adds value to patient care. The gold standard to evaluate resistance/response to therapy continues to be imaging tests. In this regard it is important to highlight that in the context of ovarian cancer, switching therapies based on increases in Ca-125 did not lead to improved survival (157,158). Also of paramount importance is to realize that not every genetic aberration found in plasma is relevant to the patient disease (see discussion in Molecular Profiling section). Fourth, turnaround times of results need to be expedited. In the MATCH study, it has taken up to 4 weeks to confirm that a genetic aberration is present. This will certainly contribute to select patients, as those who cannot wait this length of time without treatment are unlikely to be included in this study.
In summary, precision medicine is here to stay. Remarkable responses to therapy are reported across different tumor types whenever a biomarker of response is identified for which an active drug is available. We need to learn from our failures (SHIVA, BRAF inhibitors in colorectal cancer) to ensure we serve as patient advocates to identify the best available treatment options for our patients whether these are targeted therapies or not.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Katsnelson A. Momentum grows to make 'personalized' medicine more “precise”. Nat Med 2013;19:249. [Crossref] [PubMed]
- Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol 2007;3:124. [Crossref] [PubMed]
- Druker BJ. Perspectives on the development of a molecularly targeted agent. Cancer Cell 2002;1:31-6. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929-44. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine ninase, in symptomatic patients with non-small cell lung cancer: A Randomized Trial. JAMA 2003;290:2149-58. [Crossref] [PubMed]
- Rich JN, Reardon DA, Peery T, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol 2004;22:133-42. [Crossref] [PubMed]
- Garrido-Laguna I, Hidalgo M, Kurzrock R. The inverted pyramid of biomarker-driven trials. Nat Rev Clin Oncol 2011;8:562-6. [Crossref] [PubMed]
- Von Hoff DD, Stephenson JJ Jr, Rosen P, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010;28:4877-83. [Crossref] [PubMed]
- Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: The MD Anderson Cancer Center Initiative. Clin Cancer Res 2012;18:6373-83. [Crossref] [PubMed]
- Wheler J, Yelensky R, Falchook G, et al. Next generation sequencing of exceptional responders with BRAF-mutant melanoma: Implications for sensitivity and resistance. BMC Cancer 2015;15:61. [Crossref] [PubMed]
- Le Tourneau C, Delord JP, Gonçalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324-34. [Crossref] [PubMed]
- McNeil C. NCI-MATCH launch highlights new trial design in precision-medicine era. J Natl Cancer Inst 2015.107. [PubMed]
- Printz C. NCI launches exceptional responders initiative: Researchers will attempt to identify why some patients respond to treatment so much better than others. Cancer 2015;121:803-4. [Crossref] [PubMed]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet 2008;24:133-41. [Crossref] [PubMed]
- Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol 2009;12:107-18. [Crossref] [PubMed]
- Doitsidou M, Poole RJ, Sarin S, et al. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One 2010;5:e15435. [Crossref] [PubMed]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet 2010;11:31-46. [Crossref] [PubMed]
- Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet 2011;52:413-35. [Crossref] [PubMed]
- Rothberg JM, Hinz W, Rearick TM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011;475:348-52. [Crossref] [PubMed]
- Merriman B, Ion Torrent R, Team D, Rothberg JM. Progress in Ion Torrent semiconductor chip based sequencing. Electrophoresis 2012;33:3397-417. [Crossref] [PubMed]
- Singh RR, Patel KP, Routbort MJ, et al. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer 2014;111:2014-23. [Crossref] [PubMed]
- Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencingscreen for mutational hotspots in 46 cancer-related genes. J Mol Diagn 2013;15:607-22. [Crossref] [PubMed]
- Luthra R, Chen H, Roy-Chowdhuri S, et al. Next-generation sequencing in clinical molecular diagnostics of cancer: advantages and challenges. Cancers 2015;7:2023-36. [Crossref] [PubMed]
- Grosu DS, Hague L, Chelliserry M, et al. Clinical investigational studies for validation of a next-generation sequencing in vitrodiagnostic device for cystic fibrosis testing. Expert Rev Mol Diagn 2014;14:605-22. [Crossref] [PubMed]
- Strom SP, Lee H, Das K, et al. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med 2014;16:510-15. [Crossref] [PubMed]
- Park JY, Kricka LJ, Fortina P. Next-generation sequencing in the clinic. Nat Biotechnol 2013;31:990-2. [Crossref] [PubMed]
- Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet 2013;14:295-300. [Crossref] [PubMed]
- Ali SM, Alpaugh RK, Buell JK, et al. Antitumor response of an ERBB2 amplified inflammatory breast carcinoma with EGFR mutation to the EGFR-TKI erlotinib. Clin Breast Cancer 2014;14:e14-6. [Crossref] [PubMed]
- Law LY. Dramatic response to trastuzumab and paclitaxel in a patient with human epidermal growth factor receptor 2–positive metastatic cholangiocarcinoma. J Clin Oncol 2012;30:e271-3. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Schwaederle M, Husain H, Fanta PT, et al. Use of liquid biopsies in clinical oncology: pilot experience in 168 patients. Clin Cancer Res 2016;22:5497-505. [Crossref] [PubMed]
- Zill OA, Greene C, Sebisanovic D, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov 2015;5:1040-8. [Crossref] [PubMed]
- Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 2016;6:20913. [Crossref] [PubMed]
- Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015;6:7686. [Crossref] [PubMed]
- American Society of Clinical Oncology. The state of cancer care in America, 2016: A report by the American Society of Clinical Oncology. J Oncol Pract 2016;12:339-83. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A, et al. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Khan SA, Davidson BR, Goldin RD, et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 2012;61:1657-69. [Crossref] [PubMed]
- Ross JS, Wang K, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation wequencing. Oncologist 2014;19:235-42. [Crossref] [PubMed]
- Hezel AF, Deshpande V, Zhu AX. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol 2010;28:3531-40. [Crossref] [PubMed]
- Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003-10. [Crossref] [PubMed]
- Borger DR, Tanabe KK, Fan KC, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 2012;17:72-9. [Crossref] [PubMed]
- Chan-On W, Nairismägi ML, Ong CK, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet 2013;45:1474-8. [Crossref] [PubMed]
- Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. [Crossref] [PubMed]
- Rizvi S, Borad M, Patel T, et al. Cholangiocarcinoma: molecular pathways and therapeutic opportunities. Semin Liver Dis 2014;34:456-64. [Crossref] [PubMed]
- Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470-3. [Crossref] [PubMed]
- Moeini A, Sia D, Bardeesy N, et al. Molecular pathogenesis and targeted therapies for intrahepatic cholangiocarcinoma. Clin Cancer Res 2016;22:291-300. [Crossref] [PubMed]
- Goyal L, Govindan A, Sheth RA, et al. Prognosis and clinicopathologic features of patients with advanced stage isocitrate dehydrogenase (IDH) mutant and IDH wild-type intrahepatic cholangiocarcinoma. Oncologist 2015;20:1019-27. [Crossref] [PubMed]
- Saha SK, Parachoniak CA, Bardeesy N. IDH mutations in liver cell plasticity and biliary cancer. Cell Cycle 2014;13:3176-82. [Crossref] [PubMed]
- Wang P, Dong Q, Zhang C, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 2013;32:3091-100. [Crossref] [PubMed]
- Study of AG-120 in Previously Treated Advanced Cholangiocarcinoma With IDH1 Mutations (ClarIDHy) (ClarIDHy). Available online: https://clinicaltrials.gov/ct2/show/NCT02989857
- Sia D, Hoshida Y, Villanueva A, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology 2013;144:829-40. [Crossref] [PubMed]
- Ang C. Role of the fibroblast growth factor receptor axis in cholangiocarcinoma. J Gastroenterol Hepatol 2015;30:1116-22. [Crossref] [PubMed]
- Borad MJ, Champion MD, Egan JB, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet 2014;10:e1004135. [Crossref] [PubMed]
- Arai Y, Totoki Y, Hosoda F, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2014;59:1427-34. [Crossref] [PubMed]
- Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636-47. [Crossref] [PubMed]
- Tabernero J, Bahleda R, Dienstmann R, et al. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol 2015;33:3401-8. [Crossref] [PubMed]
- Javle MM, Shroff RT, Zhu A, et al. A phase 2 study of BGJ398 in patients (pts) with advanced or metastatic FGFR-altered cholangiocarcinoma (CCA) who failed or are intolerant to platinum-based. J Clin Oncol 2016;34:abstr 335.
- SEER Stat Fact: Colon and Rectum Cancer. Accessed February 13, 2016. Available online: http://seer.cancer.gov/statfacts/html/colorect.html
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008;26:374-9. [Crossref] [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [Crossref] [PubMed]
- Benson Al III, Chakravarthy A, Hamilton SR, et al. editors. Cancers of the Colon and Rectum: A Multidisciplinary Approach to Diagnosis and Management (Current Multidisciplinary Oncology). 1st edition. New York: Demos Medical Publishing, 2013.
- Asghar U, Hawkes E, Cunningham D. Predictive and prognostic biomarkers for targeted therapy in metastatic colorectal cancer. Clin Colorectal Cancer 2010;9:274-81. [Crossref] [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [Crossref] [PubMed]
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-Type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705-12. [Crossref] [PubMed]
- Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 2009;27:5924-30. [Crossref] [PubMed]
- Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 2009;101:715-21. [Crossref] [PubMed]
- Bokemeyer C, Van Cutsem E, Rougier P, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS. Eur J Cancer 2012;48:1466-75. [Crossref] [PubMed]
- Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 Trial. J Clin Oncol 2010;28:466-74. [Crossref] [PubMed]
- Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: Results from the MRC FOCUS Trial. J Clin Oncol 2009;27:5931-7. [Crossref] [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [Crossref] [PubMed]
- Kopetz S, McDonough S. S1406: Randomized phase II study of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (mCRC). J Clin Oncol 2015;33:abstrTPS790.
- Kopetz S, McDonough SL, Morris VK, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG 1406). J Clin Oncol 2017;35:abstr 520.
- Kavuri SM, Jain N, Galimi F, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov 2015;5:832-41. [Crossref] [PubMed]
- Richman SD, Southward K, Chambers P, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol 2016;238:562-70. [Crossref] [PubMed]
- Therapeutic dual inhibition of HER2 pathway in the HERACLES trial. Accessed April 11, 2016. Available online: http://www.esmo.org/Oncology-News/Therapeutic-Dual-Inhibition-of-HER2-Pathway-in-the-HERACLES-Trial
- Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 2013;3:658-73. [Crossref] [PubMed]
- Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537-40. [PubMed]
- Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69:1851-7. [Crossref] [PubMed]
- Fariña Sarasqueta A, Zeestraten EC, van Wezel T, et al. PIK3CA kinase domain mutation identifies a subgroup of stage III colon cancer patients with poor prognosis. Cell Oncol (Dordr) 2011;34:523-31. [Crossref] [PubMed]
- Mouradov D, Domingo E, Gibbs P, et al. Survival in stage III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am J Gastroenterol 2013;108:1785-93. [Crossref] [PubMed]
- Eklöf V, Wikberg ML, Edin S, et al. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer 2013;108:2153-63. [Crossref] [PubMed]
- Song L, Li Y, He B, et al. Development of Small Molecules Targeting the Wnt Signaling Pathway in Cancer Stem Cells for the Treatment of Colorectal Cancer. Clin Colorectal Cancer 2015;14:133-45. [Crossref] [PubMed]
- Hu T, Li C. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer 2010;9:236. [Crossref] [PubMed]
- Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006;25:7531-7. [Crossref] [PubMed]
- Study of WNT974 in Combination With LGX818 and Cetuximab in Patients With BRAF-mutant Metastatic Colorectal Cancer (mCRC) and Wnt Pathway Mutations. Accessed June 8, 2016. Available online: https://clinicaltrials.gov/ct2/show/NCT02278133
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43-51. [Crossref] [PubMed]
- Asaoka Y, Ijichi H, Koike K. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;373:1979. [Crossref] [PubMed]
- Jäger D, Halama N, Zörnig I, et al. Immunotherapy of colorectal cancer. Oncol Res Treat 2016;39:346-50. [Crossref] [PubMed]
- Ebert PJ, Cheung J, Yang Y, et al. MAP Kinase Inhibition Promotes T Cell and Anti-tumor Activity in Combination with PD-L1 Checkpoint Blockade. Immunity 2016;44:609-21. [Crossref] [PubMed]
- Bendell JC, Kim TW, Goh BC, et al. Clinical activity and safety of cobimetinib (cobi) and atezolizumab in colorectal cancer (CRC). J Clin Oncol 2016;34:abstr 3502.
- SEER Stat Facts: Esophageal Cancer. Accessed April 6, 2016. Available online: http://seer.cancer.gov/statfacts/html/esoph.html
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Gomez-Martin C, Plaza JC, Pazo-Cid R, et al. Level of HER2 gene amplification predicts response and overall survival in HER2-positive advanced gastric cancer treated with trastuzumab. J Clin Oncol 2013;31:4445-52. [Crossref] [PubMed]
- Janjigian YY, Werner D, Pauligk C, et al. Prognosis of metastatic gastric and gastroesophageal junction cancer by HER2 status: a European and USA International collaborative analysis. Ann Oncol 2012;23:2656-62. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Sunakawa Y, Lenz HJ. Molecular classification of gastric adenocarcinoma: Translating new insights from the Cancer Genome Atlas Research Network. Curr Treat Options Oncol 2015;16:17. [Crossref] [PubMed]
- Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 2011;43:1219-23. [Crossref] [PubMed]
- Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 2013;45:478-86. [Crossref] [PubMed]
- Wang K, Johnson A, Ali SM, et al. Comprehensive genomic profiling of advanced esophageal squamous cell carcinomas and esophageal adenocarcinomas reveals similarities and differences. Oncologist 2015;20:1132-9. [Crossref] [PubMed]
- Bang YJ, Im SA, Lee KW, et al. Randomized, Double-Blind Phase II Trial With Prospective Classification by ATM Protein Level to Evaluate the Efficacy and Tolerability of Olaparib Plus Paclitaxel in Patients With Recurrent or Metastatic Gastric Cancer. J Clin Oncol 2015;33:3858-65. [Crossref] [PubMed]
- Lee HE, Kim MA, Lee HS, et al. MET in gastric carcinomas: comparison between protein expression and gene copy number and impact on clinical outcome. Br J Cancer 2012;107:325-33. [Crossref] [PubMed]
- Lennerz JK, Kwak EL, Ackerman A, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol 2011;29:4803-10. [Crossref] [PubMed]
- Kuniyasu H, Yasui W, Kitadai Y, et al. Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem Biophys Res Commun 1992;189:227-32. [Crossref] [PubMed]
- Nakajima M, Sawada H, Yamada Y, et al. The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer 1999;85:1894-902. [Crossref] [PubMed]
- Drebber U, Baldus SE, Nolden B, et al. The overexpression of c-met as a prognostic indicator for gastric carcinoma compared to p53 and p21 nuclear accumulation. Oncol Rep 2008;19:1477-83. [PubMed]
- Smolen GA, Sordella R, Muir B, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A 2006;103:2316-21. [Crossref] [PubMed]
- Doshi S, Gisleskog PO, Zhang Y, et al. Rilotumumab exposure-response relationship in patients with advanced or metastatic gastric cancer. Clin Cancer Res 2015;21:2453-61. [Crossref] [PubMed]
- Iveson T, Donehower RC, Davidenko I, et al. Rilotumumab in combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: an open-label, dose de-escalation phase 1b study and a double-blind, randomised phase 2 study. Lancet Oncol 2014;15:1007-18. [Crossref] [PubMed]
- Catenacci DV, Henderson L, Xiao SY, et al. Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer Discov 2011;1:573-9. [Crossref] [PubMed]
- Amgen halts Rilotumumab development due to trial deaths. Accessed April 11, 2016. Published November 26, 2014. Available online: http://www.cancernetwork.com/gastrointestinal-cancer/amgen-halts-rilotumumab-development-due-increased-death-signal.
- Shah MA, Bang YJ, Lordick F, et al. METGastric: A phase III study of onartuzumab plus mFOLFOX6 in patients with metastatic HER2-negative (HER2-) and MET-positive (MET+) adenocarcinoma of the stomach or gastroesophageal junction (GEC). J Clin Oncol 2015;33:abstr 4012.
- Bachleitner-Hofmann T, Sun MY, Chen CT, et al. HER kinase activation confers resistance to MET tyrosine kinase inhibition in MET oncogene-addicted gastric cancer cells. Mol Cancer Ther 2008;7:3499-508. [Crossref] [PubMed]
- Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913-21. [Crossref] [PubMed]
- Jelic S, Sotiropoulos GC. ESMO Guidelines Working Group. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21 Suppl 5:v59-64. [Crossref] [PubMed]
- Cancer Stat Facts: Liver and Intrahepatic Bile Duct Cancer. Accessed April 11, 2016. Available online: http://seer.cancer.gov/statfacts/html/livibd.html
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Bruix J, Raoul JL, Sherman M, et al. Efficacy and safety of sorafenib in patients with advanced hepatocellular carcinoma: Subanalyses of a phase III trial. J Hepatol 2012;57:821-9. [Crossref] [PubMed]
- Villanueva A, Llovet JM. Targeted therapies for hepatocellular carcinoma. Gastroenterology 2011;140:1410-26. [Crossref] [PubMed]
- Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;389:56-66. [Crossref] [PubMed]
- Ek-Khoueiry AB, Sangro B, Yau TC, et al. Phase I/II safety and antitumor activity of nivolumab (nivo) in patients (pts) with advanced hepatocellular carcinoma (HCC): Interim analysis of the CheckMate-040 dose escalation study. J Clin Oncol 2016;34:abstr 4012.
- Hezel AF, Zhu AX. Systemic Therapy for Biliary Tract Cancers. Oncologist 2008;13:415-23. [Crossref] [PubMed]
- Chiba T, Suzuki E, Saito T, et al. Biological features and biomarkers in hepatocellular carcinoma. World J Hepatol 2015;7:2020-8. [Crossref] [PubMed]
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674-87. [Crossref] [PubMed]
- Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226-39.e4. [Crossref] [PubMed]
- Worni M, Guller U, White RR, et al. Modest improvement in overall survival for patients with metastatic pancreatic cancer: a trend analysis using the surveillance, epidemiology, and end results registry. Pancreas 2013;42:1157-63. [Crossref] [PubMed]
- Bruns CJ, Solorzano CC, Harbison MT, et al. Blockade of the epidermal growth factor receptor signaling by a novel tyrosine kinase inhibitor leads to apoptosis of endothelial cells and therapy of human pancreatic. Cancer Res 2000;60:2926-35. [PubMed]
- Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A Phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol 2007;25:1960-6. [Crossref] [PubMed]
- Kindler HL, Ioka T, Richel DJ, et al. Axitinib plus gemcitabine versus placebo plus gemcitabine in patients with advanced pancreatic adenocarcinoma: a double-blind randomised phase 3 study. Lancet Oncol 2011;12:256-62. [Crossref] [PubMed]
- Kindler HL, Niedzwiecki D, Hollis D, et al. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: Phase III Trial of the Cancer and Leukemia Group B (CALGB 80303). J Clin Oncol 2010;28:3617-22. [Crossref] [PubMed]
- Philip PA, Benedetti J, Corless CL, et al. Phase III study comparing gemcitabine plus cetuximab versus gemcitabine in patients with advanced pancreatic adenocarcinoma: Southwest Oncology Group-Directed Intergroup Trial S0205. J Clin Oncol 2010;28:3605-10. [Crossref] [PubMed]
- Van Cutsem E, Vervenne WL, Bennouna J, et al. Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol 2009;27:2231-7. [Crossref] [PubMed]
- Rougier P, Riess H, Manges R, et al. Randomised, placebo-controlled, double-blind, parallel-group phase III study evaluating aflibercept in patients receiving first-line treatment with gemcitabine for metastatic pancreatic cancer. Eur J Cancer 2013;49:2633-42. [Crossref] [PubMed]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801-6. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch A, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol 2008;26:3785-90. [Crossref] [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib Monotherapy in Patients With Advanced Cancer and a Germline BRCA1/2 Mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Kindler HL, Locker GY, Mann H, et al. POLO: A randomized phase III trial of olaparib tablets in patients with metastatic pancreatic cancer (mPC) and a germline BRCA1/2mutation (gBRCAm) who have not progressed following first-line chemotherapy. J Clin Oncol 2015;33:abtrTPS4149.
- Martínez-Bosch N, Fernández-Zapico ME, Navarro P, et al. Poly(ADP-Ribose) Polymerases. Am J Pathol 2016;186:234-41. [Crossref] [PubMed]
- Shen Y, Rehman FL, Feng Y, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res 2013;19:5003-15. [Crossref] [PubMed]
- Pishvaian MJ, Slack R, Witkiewicz A, et al. A phase II study of the PARP inhibitor ABT-888 plus temozolomide in patients with heavily pretreated, metastatic colorectal cancer. J Clin Oncol 2011;29:abstr 3502.
- Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 2009;324:217. [Crossref] [PubMed]
- Chan D, Clarke S, Gill A, et al. Pathogenic PALB2 mutation in metastatic pancreatic adenocarcinoma and neuroendocrine tumour: A case report. Mol Clin Oncol 2015;3:817-9. [PubMed]
- Chantrill LA, Nagrial AM, Watson C, et al. Precision medicine for sdvanced pancreas cancer: The individualized molecular pancreatic cancer therapy (IMPaCT) Trial. Clin Cancer Res 2015;21:2029-37. [Crossref] [PubMed]
- Tischkowitz MD, Sabbaghian N, Hamel N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology 2009;137:1183-6. [Crossref] [PubMed]
- Slater EP, Langer P, Niemczyk E, et al. PALB2 mutations in european familial pancreatic cancer families. Clin Genet 2010;78:490-4. [Crossref] [PubMed]
- Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther 2011;10:3-8. [Crossref] [PubMed]
- Pauty J, Rodrigue A, Couturier A, et al. Exploring the roles of PALB2 at the crossroads of DNA repair and cancer. Biochem J 2014;460:331-42. [Crossref] [PubMed]
- Eatrides JM, Coppola D, Al Diffalha S, et al. Microsatellite instability in pancreatic cancer. J Clin Oncol 2016;34:abstr e15753.
- Diaz LA, Uram JN, Wang H, et al. Programmed death-1 blockade in mismatch repair deficient cancer independent of tumor histology. J Clin Oncol 2016;34:abstr 3003.
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Payne SN, Maher ME, Tran NH, et al. PIK3CA mutations can initiate pancreatic tumorigenesis and are targetable with PI3K inhibitors. Oncogenesis 2015;4:e169. [Crossref] [PubMed]
- Klümpen HJ, Queiroz KC, Spek CA, et al. mTOR inhibitor treatment of pancreatic cancer in a patient with Peutz-Jeghers Syndrome. J Clin Oncol 2011;29:e150-3. [Crossref] [PubMed]
- NCT02568267 Clinical Trial. Basket Study of Entrectinib (RXDX-101) for the Treatment of Patients With Solid Tumors Harboring NTRK 1/2/3 (Trk A/B/C), ROS1, or ALK Gene Rearrangements (Fusions) (STARTRK-2). Accessed August 15, 2016. Available online: https://clinicaltrials.gov/ct2/show/NCT02568267?term=Entrectinib+STARTRK-2&rank=1
- Lindemann K, Kristensen G, Mirza MR, et al. Poor concordance between CA-125 and RECIST at the time of disease progression in patients with platinum-resistant ovarian cancer: analysis of the AURELIA trial. Ann Oncol 2016;27:1505-10. [Crossref] [PubMed]
- Rustin GJ, van der Burg ME, Griffin CL, et al. Early versus delayed treatment of relapsed ovarian cancer (MRC OV05/EORTC 55955): a randomised trial. Lancet 2010;376:1155-63. [Crossref] [PubMed]


