A KRAS wild type mutational status confers a survival advantage in pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer and is the fourth leading cause of cancer death in both men and women in the United States (1). In 2017, it is estimated that approximately 53,670 new cases will be diagnosed in the United States, and approximately 43,090 individuals will die from the disease (2). Over the last 10 years, the rates of pancreatic cancer have increased by approximately 0.5% each year (2). Despite encouraging advances in therapy, the 5-year survival rate continues to be dismal and is estimated at 8% for all stages of disease and less than 3% for distant disease.
Pancreatic cancer portends an extremely poor prognosis for many reasons. The majority of PDAC cases present at an advanced stage. The diagnostic difficulty in early stage disease is largely due to the presence of nonspecific symptoms such as fatigue, weight loss, and abdominal pain. In some cases, patients exhibit signs and symptoms related to the location of the lesion (3). Lesions in the head of the pancreas (60% to 70% of tumors) tend to be diagnosed at earlier stages because of obstructive jaundice while lesions in the body and tail (20–25% of tumors) are usually diagnosed at advanced stages (4). The only potentially curative treatment for pancreatic adenocarcinoma is surgical resection, but very few patients (15–20%) are surgical candidates at the time of diagnosis (5). The standard systemic chemotherapeutic regimens for advanced and metastatic disease include single agent gemcitabine, FOLFIRINOX, and combinations of nab-paclitaxel-gemcitabine, which have demonstrated significant clinical benefits (5). FOLFIRINOX and gemcitabine/nab-paclitaxel regimens are usually preferred in patients with appropriate performance status given their survival advantage over single agent gemcitabine as proven in phase 3 clinical trials (6).
PDAC most frequently arises from high grade pancreatic intraepithelial neoplasia (PanIN) but can also arise from cystic lesions such as intraductal papillary mucinous neoplasm (IPMN) and mucinous cystic neoplasm (MCN) (7). Microscopically, the tumor is defined as malignant epithelial cells forming glands within desmoplastic stroma (8). This desmoplastic stroma is a histologic hallmark of PDAC and consists of type 1 collagen deposition, hyaluronic acid, and proliferating myofibroblasts (pancreatic stellate cells) in a background of inflammatory cells (5). This desmoplastic stroma is typically hypovascular and under profound hydrostatic pressure. Several studies have shown that this makes chemotherapeutic drug delivery difficult (9,10).
Data suggests that the progression from premalignant pancreatic neoplasms to invasive pancreatic adenocarcinoma occurs through the successive accumulation of specific gene mutations (11). Mutations in the KRAS oncogene are considered driver mutations and are an initiating event in pancreatic ductal carcinogenesis. These have been found to be present in greater than 90% of PanIN lesions, approximately 40–65% of IPMNs, and in greater than 90% of pancreatic adenocarcinomas (12-15). While activating point mutations in codons 13 and 61 of the KRAS oncogene have been described, single nucleotide mutations in codon 12 are the most common (16). Prognostically, the presence of the G12D mutation subtype has been associated with decreased overall survival (OS) (17). In addition, the impact on PDAC biology may vary with the corrected tumor-specific allelic ratio (equal to allelic ratio divided by cellularity) and dosage of mutated KRAS. Recent reports have noted a trend for PDAC with a corrected allelic ratio of ≥10% to be associated with shorter OS (18). Additional genetic alterations in tumor suppressor genes such as cyclin-dependent kinase inhibitor 2A (CDKN2A) and TP53 lead to unrestricted cell growth and loss of apoptosis and growth arrest (19). The protein product of somatically mutated SMAD4 gene functions in the transforming growth factor beta (TGF-β) cell-signaling pathway and is associated with a poor prognosis and widely metastatic disease (20,21). Mutations in these genes are all involved in the progression from a pre-malignant lesion to invasive pancreatic adenocarcinoma (22).
Located on the short arm of chromosome 12, KRAS encodes a small GTPase that plays an active role in cellular differentiation, proliferation, migration, and apoptosis, inciting an important role in cancer signaling (23). As a result of epidermal growth factor receptor (EGFR) activation, the major downstream effector pathways of KRAS signaling are activated. These include the RAF, MEK, and mitogen-activated protein kinase (MAPK) cascade, which regulate cellular proliferation and motility. EGFR activation also results in activation of other pathways including phosphatidylinositol-3 kinase (PI3K)-AKT-mTOR cell survival cascade. This is responsible for anti-apoptotic responses and cytoskeletal organization (24). Oncogenic KRAS has been shown to alter cellular metabolism both directly and indirectly to sustain tumor growth in a hypovascular, fibrotic tumor microenvironment with extreme hypoxia and limited nutrient availability. Through KRAS-influenced metabolic reprogramming in malignant cells, macropinocytosis and autophagy allow for the acquisition of extracellular nutrients and other metabolites for energy production and unrestricted cellular proliferation (25,26). There is also an upregulation in glycolysis, mediated by mechanisms including KRAS induced expression of a lactate transporter to promote lactate efflux and prevent accumulation within the cell (27-30).
While the majority of PDACs harbor mutations in KRAS, there have been limited studies evaluating the clinical outcomes of patients with wild type KRAS PDAC. Kim et al. retrospectively studied 136 pancreatic cancer patients treated with first-line gemcitabine-based chemotherapy and found that 52.2% of their patients harbored mutations in KRAS. These patients showed a worse response to first-line gemcitabine-based chemotherapy (11.3%) compared to those with wild type KRAS (26.2%) and poorer survival (P=0.001). Additionally, this survival benefit was noted in patients with combined gemcitabine/erlotinib treatment (P=0.002) compared to gemcitabine alone (P=0.121). KRAS mutations (52.2%), independent of all other clinical parameters, demonstrated a worse outcome (31). Moreover, the impact of newer combination chemotherapy regimens across differential KRAS mutational status has not been established.
In this study, we retrospectively examined a cohort of 39 patients at our institution who were diagnosed with PDAC. Twenty-seven patients were found to harbor a mutation in KRAS (KRAS mutant) and 12 patients were wild type for the KRAS mutation (KRAS wild type). We assessed for differences in overall survival (OS) and progression free survival (PFS) for patients placed on either first-line gemcitabine-based chemotherapy or FOLFIRINOX- based therapy. Our data reveals that wild type KRAS tumors are associated with longer OS and better prognosis than mutated KRAS cases independent of the therapeutic regimen. Our data supports that the absence of a KRAS mutation in PDAC may confer a significant survival advantage.
Methods
Clinical and pathologic data collection
We examined clinical records of patients treated at the Abramson Cancer Center of the University of Pennsylvania from 2013 to 2017. Patients with a pancreatic mass (determined by clinical history and radiologic findings) and a histologic diagnosis of pancreatic or pancreaticobiliary adenocarcinoma (determined by morphology and/or immunohistochemical stains) were identified. Patients with multiple discrete primary malignancies were excluded from our analysis. A subset of patients with PDAC who underwent tumor sequencing at Penn Medicine’s Center for Personalized Diagnostics (CPD) was selected for further study. Twelve patients were identified whose tumors did not harbor KRAS mutations (KRAS wild type). Twenty-seven patients with PDAC whose tumors harbored KRAS mutations were selected as controls (KRAS mutant). Specimen sources for molecular analysis included formalin fixed-paraffin embedded (FFPE) tissue from primary pancreatectomy resections, core needle biopsies, and metastatic resections/biopsies from the lung, liver, omentum, and soft tissue. The study also included fine needle aspirate samples subjected to molecular analysis. The specimen sources for those included bile duct brushings, and primary and metastatic pancreatic adenocarcinoma lesions. Clinical data collected included age, gender, ethnicity, date of pathologic diagnosis, Eastern Cooperative Oncology Group (ECOG) performance status, serum CA19.9 levels, date of last clinic visit/date of death, OS, and treatment regimens (systemic chemotherapy and/or radiation therapy). Pathologic parameters included tumor characteristics encompassing histologic grade and TNM stage. All patients were consented in order to use their secured personal data and clinical information for the purposes of the study. The study was approved by the Institutional Review Board at the University Pennsylvania prior to the collection of data (approval # 827031). The study outcomes did not affect future management of patients.
Specimen selection and next generation sequencing
Forty specimens from 39 patients were analyzed at the CPD. FFPE sections were examined by surgical pathologists for adequacy. The specimen was determined to be adequate for DNA extraction if the tumor volume was estimated to be greater than 10% at histologic diagnosis. Enrichment for tumor was performed via macro-dissection. Adequate specimens were run on the CPD Comprehensive Solid Tumor sequencing panels to detect mutations (see genes analyzed in Supplementary). Specimens with insufficient DNA yield for analysis on the full gene panel were run on the Penn Precision Panel (PPP), a limited panel of twenty genes (see genes analyzed Supplementary).
Briefly, genomic DNA was extracted from FFPE tissue or fine needle aspirates according to manufacturer’s instructions (Qiagen, Inc.). Targeted analysis for mutations on the two versions of the Solid Tumor panel was achieved by enrichment of specific genomic loci using either the Illumina TruSeq Amplicon assay that targets 47 cancer related genes (Illumina Inc., San Diego, CA, USA) or a custom Agilent Haloplex assay encompassing 153 genes (Agilent, Santa Clara, CA, USA). Sequencing of enriched libraries was performed on the Illumina MiSeq and HiSeq platforms using multiplexed, paired end reads. Analysis and interpretation utilized a customized bioinformatics process, and variant classifications were made using the hg19 Genome build (32). Targeted analysis on the PPP was achieved by enrichment of 77 amplicons enriched for hotspots and tumor suppressor coverage of 20 genes. PCR-enriched libraries were created and sequencing was performed on the Illumina MiSeq platform using multiplexed, paired end reads.
Mutations were classified as pathogenic, variants of uncertain significance, or benign. This determination was based on a literature review, and the existence of each mutation in publically available databases including dbSNP, COSMIC, ExAC, and the 1000 genome project. Pathogenic variants were defined as those with known or predicted loss or gain of function of the protein products. Variants of undetermined significance were excluded from further analysis in the study.
A subset of tumors was subject to BRCA1, and BRCA2 mutational analysis (Illumina MiSeq platform) and immunohistochemistry for the mismatch repair (MMR) proteins (MLH1, MSH2, PMS2 and MSH6).
Statistical analysis and outcome measures
The relationship between KRAS mutational status and clinical outcome was studied. OS was calculated from the date of pathologic diagnosis to the date of death or the date of the last confirmed contact with our medical system. Clinical PFS data was gathered from the patient’s medical records based on outpatient visit notes. For mean OS, 95% confidence intervals and P values were calculated via a two-sample t-test. For median OS, values were compared using Mood’s median test with Yate’s correction of the Chi square test. Survival curves were generated using the Kaplan-Meier method with living patients censored at the time of their last contact with the medical system. Survival curves were assessed for statistically significant differences via the log rank test. Statistical significance was assumed at P<0.05. This analysis was performed using Excel (v.2016, Microsoft, Redmond, WA, USA) and the Real Statistics Resource Pack (v.4.14, Real Statistics, Trento, Italy).
Results
Patient characteristics
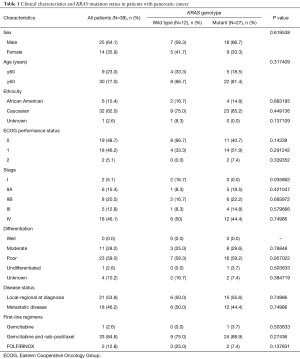
Tumor samples from a total of 39 patients were included in the study. Twenty-five patients (64.1%) were male and 30 patients (77.0%) were over the age of 60 years. There were 6 African American patients (15.4%) and 32 Caucasian patients (82.0%). Twenty-one patients (53.8%) had local-regional disease at diagnosis compared to 18 patients (46.2%) with metastatic disease. The majority of patients (94.9%) were listed as having an ECOG status of 0 or 1. Thirty-four patients (87.2%) were treated with either gemcitabine alone or gemcitabine with nab-paclitaxel as first-line chemotherapy. Five patients (12.8%) were administered a FOLFIRINOX-based first-line chemotherapeutic regimen. Patient characteristics are summarized in Table 1.
Full table
Frequency of KRAS mutations
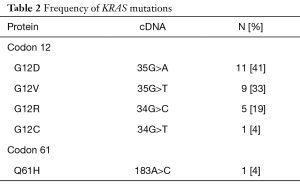
In this study, 12 of the 39 patients (31%) were found to have PDAC with a non-mutated KRAS gene (wild type). Twenty-seven of the 39 patients (69%) were found to have tumors with a mutated KRAS gene. In the KRAS mutant group, the most common single nucleotide mutation occurred at codon 12 (Table 2). Observed point mutations at codon 12 in the order of frequency were: (G12D) GGT-GAT (35G>A; 11 of 27 patients, 41%); (G12V) GGT-GTT (35G>T; 9 of 27 patients, 33%); (G12R) GGT-CGT (34G>C; 5 of 27 patients, 19%); (G12C) GGT-TGT (34G>T; 1 of 27 patients, 4%). There were no mutations identified in codon 13 and only 1 mutation was detected in codon 61: (Q61H) CAA-CAC (183A>C; 1 of 27 patients, 4%). 16.7% of KRAS wild type patients in our study presented with stage 1 disease at diagnosis compared to only 5.1% of KRAS mutant patients (P=0.03). However, other clinical and pathologic parameters such as sex, age, ethnicity, ECOG performance status, histologic differentiation, or disease status were not associated with KRAS mutations.
Full table
MMR status, BRCA, and other gene mutations
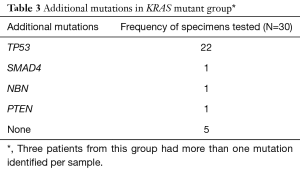
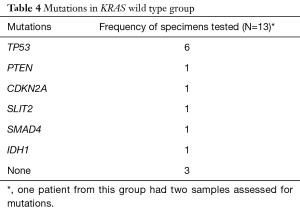
TP53 was the most common additional mutation noted in both the KRAS mutant (22 of 27, 81.5%) and KRAS wild type (6 of 12, 50%) patients (Tables 3,4). When possible, MMR status was assessed by immunohistochemical stains for MLH1, PMS2, MSH2, and MSH6 [17 of 39 patients (44%), 12 patients with KRAS mutant tumors and 5 patients with KRAS wild type tumors, data not shown]. All tested tumor cases were MMR proficient. Additionally, 7 of 39 patients (17.9%, 2 KRAS wild type and 5 KRAS mutant, data not shown), were tested for BRCA1 and BRCA2 mutations. No mutations in either gene were identified in our cohort.
Full table
Full table
Impact of KRAS mutation on survival
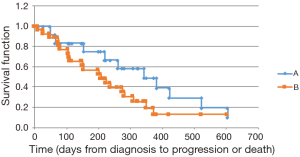
We noted a longer OS among KRAS wild type patients [mutant KRAS, observed median survival of 402 days (mean 420.6 days, 95% CI: 335.1–505.9) vs. wild type KRAS, observed median of 720 days (mean 763.6 days, 95% CI: 579.3–947.9); median P=0.011, mean P=0.005]. Similar findings were obtained via the Kaplan-Meier survival analysis (mutant KRAS, calculated median OS of 531 days vs. wild type KRAS, median 957 days; P=0.026) (Figure 1). An analysis of PFS showed a trend toward a longer PFS in the wild type group; however, this finding did not achieve statistical significance (mutant KRAS, calculated median PFS of 208 days vs. wild type KRAS, median 343 days; P=0.247) (Figure 2). Thirty-four of the 39 patients (87%) received first-line gemcitabine-based chemotherapy (gemcitabine alone or in conjunction with nab-paclitaxel), and 5 patients received FOLFIRINOX therapy. The therapeutic regimens were determined independent of the tumor mutational status. No significant difference was discerned in response to therapy between the two groups.

Pathologic features
Histomorphologic features were assessed for the KRAS wild type and KRAS mutant cases. There was no significant difference in the grade of tumor differentiation of the pancreatic adenocarcinoma between the two groups. Of the KRAS wild type cases, 25% were moderately differentiated and 58.3% were poorly differentiated, compared to 29.6% and 59.2% of KRAS mutant cases, respectively. Tumors from the KRAS mutant group demonstrated variant morphologic features. Of the four cases available for review in this group, two showed a syncytial growth pattern with pushing borders and without well-formed glands. One of the tumors demonstrated a microglandular pattern with luminal necrosis. The remaining two KRAS wild type tumors demonstrated conventional morphology with infiltrating poorly formed glands (Figure 3).

Discussion
It is well known that activating point mutations in the KRAS oncogene are present in a significant percentage of precursor lesions such as PanIN as well as in PDAC (12,14). As a downstream effector in the EGFR pathway, KRAS encodes a small (approximately 21kDa) GTPase, P21 RAS, which cycles between an active (GTP bound) and inactive (GDP bound) state and influences various intracellular signaling pathways (33).Mutations in KRAS cause constitutive activation of P21 RAS and perpetuate downstream signaling pathways involved in cellular proliferation, migration, apoptosis, and cytoskeletal remodeling, independent of growth factor receptor activation (33).
Previous studies have attempted to characterize the role of KRAS as a prognostic biomarker for clinical outcomes in PDAC. As previously mentioned, Kim et al. demonstrated that KRAS wild type patients showed a better objective response, longer OS, and a lower risk of death compared to KRAS mutant patients when treated with first-line gemcitabine-based chemotherapeutic regimens. On combination chemotherapy regimens with gemcitabine and erlotinib (an EGFR tyrosine kinase inhibitor), wild type KRAS patients were found to have longer median survivals. Similar results were shown in a different study analyzing outcomes for patients with KRAS mutant and KRAS wild type PDAC on erlotinib combined with either gemcitabine or capecitabine (34). A recent German study compared the 12-month OS of patients with advanced PDAC treated with gemcitabine and nimotuzumab (an anti-EGFR monoclonal antibody) as first line therapy versus gemcitabine plus placebo, and found that KRAS wild type patients had a significant survival benefit compared to those with KRAS mutations (11.6 versus 5.6 months, P=0.03) (35). While most studies have suggested a survival advantage for patients with KRAS wild type PDAC, this finding has not been universal. In a multicenter prospective study from Bournet et al., tumor from 219 patients, obtained via ultrasound-guided fine needle aspiration biopsies was sequenced for KRAS mutations. No significant difference in OS was noted between the subtypes of mutant KRAS (G12D, G12V, and G12R) and wild type KRAS samples.
In our single institution retrospective study, we investigated the role of KRAS mutations on the prognosis of patients with PDAC. We observed an increased OS in patients with wild type KRAS tumors compared to those with mutant KRAS. We also noted a trend toward increased PFS in the wild type patients, however, this result did not achieve statistical significance. This is most likely due to the small sample size, reflecting the limited power of our study. In contrast to previous studies, which demonstrated a difference in survival based on the type of gemcitabine combination therapy and KRAS mutational status, these differences were not appreciated within our limited cohort. Our study also did not show any appreciable difference in survival between KRAS wild type and mutant patients treated with a specific gemcitabine-based first-line therapy. Moreover, within our study group, five patients received FOLFIRINOX first-line therapy, and there was no difference in survival within these patients. Similar to previous studies, we demonstrated that KRAS mutations were not associated with other clinical parameters including sex, age, and ECOG performance status (31).
The replacement of nucleotide sequence GGT with GAT in codon 12 of exon 2 of the KRAS gene (G12D subtype) has been described as the most frequently observed mutation in PDAC (13,17). It has also been shown to be a negative prognostic indicator for OS in patients treated with or without first-line gemcitabine chemotherapy (17). Similar to previous studies, the KRAS G12D subtype was the most frequently observed point mutation in our cohort. Mutations in codons 13 and 61 of KRAS are more common in colorectal cancer (15% and 1%, respectively) but have been rarely reported in PDAC (36,37). Our study population showed no mutations in codon 13 and only 1 mutation in codon 61. No relationship was appreciated between specific activating point mutations with KRAS and survival outcomes or responses to chemotherapy.
In addition to KRAS mutations, it is well documented that somatic gene mutations in p16/CDKN2A, TP53, and SMAD4/DPC4 are also noted in advanced pancreatic cancer. The presence of additional pathogenic mutations within each group was assessed. While loss of function of the tumor suppressor gene, CDKN2A/p16, has been reported in approximately 90% of pancreatic cancers (38), our study found that mutations in the tumor suppressor gene, TP53, were the most common associated mutation irrespective of the KRAS mutational status. Inactivation of TP53 can affect cell cycle regulation of the G1-S checkpoint, G2-M arrest, and apoptosis. Some studies have reported that mutations in TP53 are seen in as many as 50–76% of pancreatic cancers (39). Results from our study were similar to those reported in the literature. We found mutations in TP53 in 81% (22/27) of tumor samples in the mutant group and in 46% (6/13) of tumor samples in the wild type group. No CDKN2A mutations were detected in the KRAS mutant group, and only one concurrent CDKN2A mutation was seen in the KRAS wild type group.
Numerous studies have assessed the features of carcinomas with errors in DNA replication. It is well understood that germline and somatic mutations in the DNA MMR genes hMLH1, hMSH2, hMSH6, and hPMS2 confer a higher lifetime risk of developing pancreatic cancer. All tested KRAS wild type and mutant samples were MMR proficient. Inherited mutations in other DNA repair genes such as BRCA2 and less commonly in BRCA1 (40-43) have also been associated with an elevated risk of pancreatic cancer; no mutations were detected in the BRCA1 and BRCA2 genes in our study.
Some studies have shown a correlation between the KRAS mutational profile and unique phenotypic characteristics of the tumor. The presence of a medullary tumor morphology (poor differentiation, expanding borders, and a syncytial growth pattern) in PDAC has been associated with a wild type KRAS mutational status and/or microsatellite instability (44,45). MMR testing was done on only 5 or 12 patients of the KRAS wild type group; all of these patients were found to be proficient for the MMR proteins. Histologic features were assessed for four of the twelve patients in this group. Two of these demonstrated a syncytial growth pattern with pushing borders, while the other two showed a conventional infiltrative growth pattern. Therefore, although our sample size was small, we did not find any association between KRAS wild type mutational status and morphologic features. Kim et al. demonstrated that a moderately differentiated histology was a prognosticator for better OS in PDAC. We did not find any correlation between tumor differentiation (well, moderate, poor, and undifferentiated) and mutation status in our study.
Overall, similar to previously reported studies, PDACs with a KRAS wild type mutational profile have a better prognosis with a longer OS. This improved prognosis is independent of the protocol utilized in therapy for these patients. These findings suggest that future clinical trials in pancreatic cancer should take into consideration the presence of KRAS mutations in their pre-planned analysis when assessing the efficacy of a novel therapeutic approach. This may be a crucial factor in trial concepts and outcomes.
Supplementary
Genes included in Comprehensive Solid Tumor Panel v1 (cases from 2013 to early 2016): ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL.
Genes included in Comprehensive Solid Tumor Panel v2 (implemented in late 2016): ABL1, AKT1, AKT2, AKT3, ALK, APC, AR, ARAF, ARID1A, ARID2, ATM, ATRX, AURKA, BAP1, BRAF, BRCA1, BRCA2, BRIP, BTK, CBP, CCND1, CCND2, CCND3, CCNE1, CDH1, CDK4, CDK6, CDKN2A, CHEK2, CIC, CRKL, CSF1R, CTNNB1, DAXX, DDR2, DNMT3A, EIF1Ax, EGFR, EP300, EPHA3, ERBB2, ERBB3, ERBB4, ERCC2, ERG, ESR1, ESR2, EZH2, FBXW7, FGF3, FGFR1, FGFR2, FGFR3, FGFR4, FLT3, FUBP1, GATA3, GNA11, GNAQ, GNAS, HRAS, H3F3A, IDH1, IDH2, IGF1R, JAK1, JAK2, JAK3, KCNG1, KDM5A, KDM5C, KDM6A, KDR, KIT, KMT2C, KRAS, MAP2K1, MAP2K2, MAP2K4, MAPK1, MAPK3, MAX, MCL1, MDM2, MDM4, MED12, MEN1, MET, MITF, MLH1, MRE11A, MSH2, MSH6, MTOR, MYC, MYCN, NBN, NF1, NF2, NKRT1, NKRT2, NKRT3, NKX2-1, NOTCH1, NOTCH2, NOTCH3, NRAS, PAK1, PALB2, PBRM1, PDGFRA, PIK3CA, PIK3CB, PIK3R1, PTCH1, PTEN, PTPN11, RAB35, RAC1, RAD50, RAD51, RAD51B, RAD51C, RAD51D, RAF1, RB1, RET, RHOA, RNF43, SETD2, SF3B1, SLIT2, SMAD4, SMARCA4, SMO, SPOP, SRC, STAG2, STK11, SUFU, SUZ12, SYK, TET2, TGFBR2, TP53, TRAF7, TSC1, TSHR, TSC2, U2AF1, VHL, WT1, XRCC2.
Genes included in Penn Precision Panel: AKT1, ALK, BRAF, CS1R, EGFR, ERBB2, HRAS, IDH1, IDH2, KIT, KRAS, MAP2K1, MET, NOTCH1, NRAS, PDGRRA, PIK3CA, PTEN, RET, TP53.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Institutional Review Board at the University Pennsylvania prior to the collection of data (approval # 827031). All patients were consented in order to use their secured personal data and clinical information for the purposes of the study.
References
- Society AC. Cancer Facts and Figures 2017. Atlanta: American Cancer Society, 2017.
- Surveillance, Epidemiology, and End Results Program (SEER) Cancer Stat Facts: Pancreas Cancer. National Cancer Institute. Available online: https://seer.cancer.gov/statfacts/html/pancreas.html
- Modolell I, Guarner L, Malagelada JR. Vagaries of clinical presentation of pancreatic and biliary tract cancer. Ann Oncol 1999;10 Suppl 4:82-4. [Crossref] [PubMed]
- Ducreux M, Cuhna AS, Caramella C, et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2015;26 Suppl 5:v56-68. [Crossref] [PubMed]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:2140-1. [Crossref] [PubMed]
- Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers 2016;2:16022. [Crossref] [PubMed]
- Matthaei H, Schulick RD, Hruban RH, et al. Cystic precursors to invasive pancreatic cancer. Nat Rev Gastroenterol Hepatol 2011;8:141-50. [Crossref] [PubMed]
- Hruban RH KD, Pitman MB. Atlas of Tumor Pathology. Tumors of the pancreas. Fourth Series Edition ed. Washington, DC: Armed Forces Institute of Pathology, 2006.
- Provenzano PP, Cuevas C, Chang AE, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 2012;21:418-29. [Crossref] [PubMed]
- Jacobetz MA, Chan DS, Neesse A, et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut 2013;62:112-20. [Crossref] [PubMed]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004;10:789-99. [Crossref] [PubMed]
- Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012;142:730-3. e9.
- Hruban RH, van Mansfeld AD, Offerhaus GJ, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol 1993;143:545-54. [PubMed]
- Hong SM, Vincent A, Kanda M, et al. Genome-wide somatic copy number alterations in low-grade PanINs and IPMNs from individuals with a family history of pancreatic cancer. Clin Cancer Res 2012;18:4303-12. [Crossref] [PubMed]
- Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med 2011;3:92ra66. [Crossref] [PubMed]
- Ying H, Dey P, Yao W, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2016;30:355-85. [Crossref] [PubMed]
- Bournet B, Muscari F, Buscail C, et al. KRAS G12D Mutation Subtype Is A Prognostic Factor for Advanced Pancreatic Adenocarcinoma. Clin Transl Gastroenterol 2016;7:e157. [Crossref] [PubMed]
- Lennerz JK, Stenzinger A. Allelic ratio of KRAS mutations in pancreatic cancer. Oncologist 2015;20:e8-9. [Crossref] [PubMed]
- Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin 2013;63:318-48. [Crossref] [PubMed]
- Iacobuzio-Donahue CA, Fu B, Yachida S, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol 2009;27:1806-13. [Crossref] [PubMed]
- Blackford A, Serrano OK, Wolfgang CL, et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res 2009;15:4674-9. [Crossref] [PubMed]
- Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet 2011;378:607-20. [Crossref] [PubMed]
- Cox AD, Der CJ. Ras history: The saga continues. Small GTPases 2010;1:2-27. [Crossref] [PubMed]
- Normanno N, Tejpar S, Morgillo F, et al. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol 2009;6:519-27. [Crossref] [PubMed]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013;497:633-7. [Crossref] [PubMed]
- Yang S, Wang X, Contino G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 2011;25:717-29. [Crossref] [PubMed]
- Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012;149:656-70. [Crossref] [PubMed]
- Racker E, Resnick RJ, Feldman R. Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc Natl Acad Sci U S A 1985;82:3535-8. [Crossref] [PubMed]
- Yun J, Rago C, Cheong I, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 2009;325:1555-9. [Crossref] [PubMed]
- Baek G, Tse YF, Hu Z, et al. MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep 2014;9:2233-49. [Crossref] [PubMed]
- Kim ST, Lim DH, Jang KT, et al. Impact of KRAS mutations on clinical outcomes in pancreatic cancer patients treated with first-line gemcitabine-based chemotherapy. Mol Cancer Ther 2011;10:1993-9. [Crossref] [PubMed]
- Daber R, Sukhadia S, Morrissette JJ. Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet 2013;206:441-8. [Crossref] [PubMed]
- Hingorani SR, Tuveson DA. Ras redux: rethinking how and where Ras acts. Curr Opin Genet Dev 2003;13:6-13. [Crossref] [PubMed]
- Boeck SH, Vehling-Kaiser U, Waldschmidt D, et al. Gemcitabine plus erlotinib (GE) followed by capecitabine (C) versus capecitabine plus erlotinib (CE) followed by gemcitabine (G) in advanced pancreatic cancer (APC): a randomized, cross-over phase III trial of the Arbeitsgemeinschaft Internistische Onkologie (AIO). J Clin Oncol 2010;28:abstr LBA4011.
- Schultheis B, Reuter D, Ebert MP, et al. Gemcitabine combined with the monoclonal antibody nimotuzumab is an active first-line regimen in KRAS wildtype patients with locally advanced or metastatic pancreatic cancer: a multicenter, randomized phase IIb study. Ann Oncol 2017;28:2429-35. [Crossref] [PubMed]
- Edkins S, O'Meara S, Parker A, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther 2006;5:928-32. [Crossref] [PubMed]
- Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol 2012;18:5171-80. [PubMed]
- Maitra A, Hruban RH. Pancreatic Cancer. Annu Rev Pathol 2008;3:157-88. [Crossref] [PubMed]
- Redston MS, Caldas C, Seymour AB, et al. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res 1994;54:3025-33. [PubMed]
- Goggins M, Schutte M, Lu J, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res 1996;56:5360-4. [PubMed]
- Ferrone CR, Levine DA, Tang LH, et al. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J Clin Oncol 2009;27:433-8. [Crossref] [PubMed]
- Thompson D, Easton DF. Breast Cancer Linkage Consortium. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst 2002;94:1358-65. [Crossref] [PubMed]
- Axilbund JE, Argani P, Kamiyama M, et al. Absence of germline BRCA1 mutations in familial pancreatic cancer patients. Cancer Biol Ther 2009;8:131-5. [Crossref] [PubMed]
- Goggins M, Offerhaus GJ, Hilgers W, et al. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+. Am J Pathol 1998;152:1501-7. [PubMed]
- Wilentz RE, Goggins M, Redston M, et al. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am J Pathol 2000;156:1641-51. [Crossref] [PubMed]


