Evaluation of altered expression of miR-9 and miR-106a as an early diagnostic approach in gastric cancer
Introduction
Gastric cancer (GC) is the fourth common malignancy and the second cause of cancer-related deaths in the world as well as the first cause of cancer mortality in Iranian population. Although lung and breast cancers are very common among males and females, respectively, but, digestive system cancers such as esophagus, gastric, liver, colon, and pancreas are responsible for three millions of new cases and more than two millions of mortality per year (1). Lack of a specific diagnostic symptom in early stages, non-efficiency of existing diagnostic approaches such as endoscopy imaging methods (2), weak prognosis of GC (79% of tumors diagnosed in the fourth stage of disease and less than 5% of patients have life expectancy of 5 years), and chemotherapy resistance in advanced stages will encounter diagnosing and treating of this cancer with complicity. However, if this cancer is diagnosed in its early stages, the treatment will be more successful (3). For reasons mentioned above, it is necessary to have a desirable diagnostic method. Recently, cancer researchers have focused on biomarkers. Biomarkers are biological molecules found in blood and other body fluids or tissues and can be a sign of normality or abnormality of internal conditions of the body (4). Among existing biomarkers, miRNA seems to be more suitable for diagnostic and treatment objectives. Moreover, they have been used in cancer researches and it is expected to improve the diagnosis and treatment of cancers (5). miRNAs are non-coding RNAs with 18–25 nucleotides which are protected during evolution (6) and are involved in cellular processes such as cell cycle regulation, differentiation, stress response, inflammation, apoptosis, and migration. Thus, miRNAs have been implicated in almost all signal transduction pathways within a cell and their dysregulation plays an essential role in cancer development (7). These molecules bind to the non-coding region in 3'UTR of target mRNAs and control the gene expression via the translation inhibition or degradation of target mRNA (6). This study tries to investigate the expression rate of miR-106a which its role in carcinogenesis has been proved in several studies (8,9) and miR-9 which there are some challenges related to its role in carcinogenesis (10-13) in GC tissues in comparison with healthy adjacent tissues.
Methods
Patients and specimens
This is a case-control study conducted on 31 tumorous samples of GC patients in the age group of 31–83 years who did not receive any treatment. Since the aim of this study was an early diagnosis of cancer, samples that were in their first or second stages with no advanced metastasis were included. The disease stage was determined by a taking biopsy during endoscopy and verified by a pathologist. Samples were prepared based on ethical principles and an informed consent was taken from the patients (previously taken by the staff of Imam Khomeini Hospital). The healthy adjacent tissues of the same patients were used as control group. The healthy adjacent tissues were farther than 5 cm from the tumor and there were no tumorous cells, as evaluated by a pathologist.
RNA extraction
In order to conduct the test, extracting total RNAs from tumorous and healthy tissues were required. For this purpose, all prepared tissues were crushed by a homogenizer. For disrupting cells and dissolving cell components Trizol (Invitrogen, USA) was added according to manufacturer’s instruction. In the next stage, chloroform was added and the sample was centrifuged at 12,000 ×g for 15 minutes at 4 °C. The supernatant containing RNA was isolated and placed into a new tube and the same volume of isopropanol was added. The obtained mixture was incubated at room temperature for 10 minutes and centrifuged with in the previous conditions. Once more, the supernatant was removed and 1 mL ethanol 75% was added to the remaining RNA pellet and then centrifuged at 7,500 ×g for 5 minutes at 4 °C. Next, the alcohol was discarded and RNA pellet was dried at 55 °C for 10 min. RNA concentration and purity were controlled by NanoDrop Spectrophotometer (Biotek EPOCH, USA). Finally, RNA pellet was resuspended in RNase-free water and stored in −80 °C.
Measurement of miRNA expression
Real time PCR processes were done by ParsGenome’s miRNA amplification Kit based on the guidelines of the manufacturer as below:
Poly A polymerase enzyme addition
1.5 µg of RNA was added to 2 µL buffer 10X, 1 µL ATP (10 mM), 0.5 µL Poly A enzyme and DEPC-treated water and then incubated at 37 °C for 10 min.
First-strand cDNA synthesis
6 µL of obtained poly delineated RNA was mixed in 2 µL buffer 5X, 0.5 µL RT enzyme as well as 0.5 µL miRNA cDNA synthesis specific primer (15 pmol) and incubated at 42 °C for 15 min. For inactivating RT enzyme the mixture was stored at 85 °C for 15 min.
Real-time PCR amplification
10 µL SYBR Green master mix, 1 µL miR specific primers (10 pmol, designed by Pars Genome Company), and 1 µg of diluted cDNA were mixed together. The thermal cycling conditions included: 5 minute at 95 °C, 5 seconds at 95 °C, 20 seconds at 62 °C, and 30 seconds at 72 °C. Thermal cycling proceeded with 35 cycles. No template control (NTC) was used for controlling the contamination (14). Moreover, for data normalization 5srRNA was used (15).
Statistical analysis
In order to determine the expression rate differences of the miRNAs in tumorous and healthy adjacent tissues the averages of ΔCt (CTmiRNA − CT5srRNA) were compared using paired sample t-test and independent sample t-test was used for statistical analyses of miRNAs expression rate differences in different ages, genders, and stages. The “fold change” was calculated by 2-ΔΔCT formula. ΔΔCT = (CTmiRNA − CT5srRNA) tumorous tissues − (CTmiRNA − CT5srRNA) healthy adjacent tissues (14,16,17).
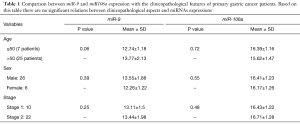
The clinicopathological features of 31 GC patients are shown in Table 1. miR-106a and miR-9 expression had no significant relationship with clinicopathological parameters (age, gender, stage).
Full table
Results
Expression of miR-9 in tumorous and healthy adjacent tissues
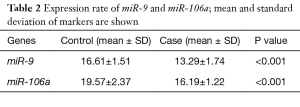
The expression rate of miR-9 decreased in 3.33% cases (one sample) and increased in 96.77% cases (30 samples). This miRNA shows a significant expression difference between both groups (tumorous and healthy adjacent tissues) (Table 2). The expression fold change of miR-9 was 10.41, which means that the expression rate of miR-9 increased 10.41-fold in the tumorous tissues (Table 3).
Full table

Full table
Expression of miR-106a in tumorous and healthy adjacent tissues
The expression rate of miR-106a increased in 96.77% cases and statistical analysis shows that miR-106a expression was significantly different between control (healthy adjacent) and case (tumorous) groups (P<0.001) (Table 2). The expression fold change of miR-106a was 10.33, which means that the expression rate of miR-106a increased 10.33-fold in the tumorous tissues (Table 3).
Receiver operating characteristic (ROC) curve analysis
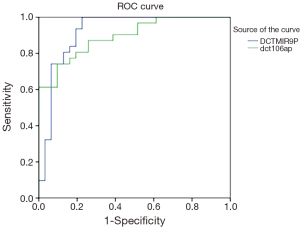
ROC curve analyses were accomplished in order to determine the specificity and sensitivity of these miRNAs as diagnostic biomarkers (18).
The area under the curve (AUC or c-statistic or c index) can range from 0.5 to 1 (no predictive ability to perfect differentiation) (19). The results indicated that AUC for miR-9 equal 0.919 and for miR-106a equal 0.894, which reveals the ability of these miRNAs as diagnostic biomarkers are very high (Figure 1).
Discussion
GC is a polygenic disease, meaning that several genes play roles in cancer development and progression, and during several stages of cancer their expression will be dysregulated. However, the accurate mechanism of this process is unknown (10).
MicroRNAs (miRNAs) expression has been altered in several cancer types and may have a causative role (oncogenic or tumor-suppressive role) in cancer development. The prognostic capacity of miRNAs has been described for different cancers (17).
Improving therapeutic and particularly diagnosis approaches in the early stages of GC may increase the life expectancy and survival of patients, while delay in diagnosis and treatment leads to reduced survival rate in patients. Due to the absence of proper approach in addition to sensitive and specific biomarker, more research is required in order to find suitable biomarkers (20).
This study has investigated the expression rate of miR-9 and miR-106a. The results indicate that the expression rate of both miRNAs increased in the tumorous tissues. Moreover, the results of this study confirm the results of previous studies and show that reduced expression rate of miR-106a in one sample out of 31 examined samples, which this means that the expression of this miRNA increased in 96.77%. In addition expression differences between both groups was significant (P<0.001). In a study conducted by Tsujiura et al. increased expression of miR-106a in the blood samples of GC patients have been reported (P=0.008) (21). In study on the tumorous tissue which was conducted by Yao, et al. upregulation of miR-106a (2.83-fold) was reported (P=0.03) (22). Also, Guo et al. showed that the expression rate of miR-106a significantly increased in the GC tissues and this upregulation was correlated with reduced expression of Rb tumor suppressor (23). The calculated fold change by Xiao et al. and Li et al. for miR-106a in the GC sample was respectively 1.625 and 5.98 (17,24) and in the current study, it was calculated 10.33. Comparing such results to previous studies indicates a possible relation between this miRNA and GC. Moreover, in a study done by Konishi et al. the reported sensitivity for miR-106a in plasma and serum was respectively 85% and 48.2% (25). In the current study, the obtained sensitivity in gastric tissue was 96.77% and this sensitivity is very considerable. On other hand, in contrast to some previous studies, in the present study there is no relationship between this miRNA expression, age, and gender which means that these factors have no influence on the expression rate of miR-106a and therefore, results could be interpreted by more confidence. Contrary to Xiao study (17), statistical analysis of this study indicated that there is no significant relation between expression rate of miR-106a and the stage of GC.
Statistical analysis indicates significant expression rate difference of miR-9 between both groups (P<0.001). miR-9 expression rate was increased in 96.77% of tumorous samples and its fold change compared with healthy adjacent tissues which was 10.41. In comparison to our findings, Rotkrua et al. indicated reduced expression rate of miR-9 in 73% tumorous samples (P=0.0073) (26).
The most performed studies on miR-9 are about evaluation of aberrant methylation of miR-9 promoter CpG-island. Tsai et al. have indicated that miR-9 can function as a tumor suppressor and they observed high frequency of hypermethylation at three primary miR-9 transcripts in GC cells, that resulted in miR-9 downregulation in GC (12). In contrast, Rotkrua et al. showed that methylation of this miRNA is significantly correlated with the increased expression rate of CDX2 protein. In addition, they indicated that the expression level of CDX2 was significantly downregulated in miR-9-transfected cells. Also, they revealed the tumor-suppressive role of CDX2 and oncogenic role of miR-9 in GC (26). By the way in current study, upregulation of miR-9 in tumorous tissue confirmed the oncogenic role of miR-9.
In the current study the AUC value for miR-9 was obtained 0.919 and for miR-106a it was 0.894, indicating a very good ability of these miRNAs for differentiating tumorous and healthy samples. Unfortunately, in the previous studies the results from ROC curve, by which the results can be compared, have not been stated.
Conclusions
In conclusion, it is complicated to judge about the role of miR-9 in carcinogenesis. Therefore, more studies are required and their results must be interpreted with more accuracy. The objectives of this marker and its mechanism of action require more studies. This miRNA might not be a suitable biomarker for diagnosis of GC and its results are not reliable. However, regarding miR-106a with high confidence, it could be considered as a suitable biomarker. Results obtained from this study are more acceptable comparing to previous results obtained from this miRNA. Therefore, miR-106a is more likely to be one of the genes involved in carcinogenesis.
Acknowledgements
This project was financially supported by the Research and Technology Deputy of Hamadan University of Medical Sciences and was a part of MSc thesis of K Shirmohammadi.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by Medical Ethical Committee of Hamadan University of Medical Sciences(ID: D/P/16/35/9/314) and informed consent was taken from all the patients.
References
- Sadjadi A, Malekzadeh R, Derakhshan MH, et al. Cancer occurrence in Ardabil: results of a population-based cancer registry from Iran. Int J Cancer 2003;107:113-8. [Crossref] [PubMed]
- Dhalla F, da Silva SP, Lucas M, et al. Review of gastric cancer risk factors in patients with common variable immunodeficiency disorders, resulting in a proposal for a surveillance programme. Clin Exp Immunol 2011;165:1-7. [Crossref] [PubMed]
- Cooke CL, Torres J, Solnick JV. Biomarkers of Helicobacter pylori-associated gastric cancer. Gut microbes 2013;4:532-40. [Crossref] [PubMed]
- Yin Y, Li J, Chen S, et al. MicroRNAs as diagnostic biomarkers in gastric cancer. Int J Mol Sci 2012;13:12544-55. [Crossref] [PubMed]
- Gao M, Yin H, Fei ZW. Clinical application of microRNA in gastric cancer in Eastern Asian area. World J Gastroenterol 2013;19:2019-27. [Crossref] [PubMed]
- Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol 2008;18:103-10. [Crossref] [PubMed]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [Crossref] [PubMed]
- Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med 2006;12:580-7. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Luo H, Zhang H, Zhang Z, et al. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res 2009;28:82. [Crossref] [PubMed]
- Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene 2006;25:6170-5. [Crossref] [PubMed]
- Tsai KW, Liao YL, Wu CW, et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics 2011;6:1189-97. [Crossref] [PubMed]
- Zhu L, Chen H, Zhou D, et al. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol 2012;29:1037-43. [Crossref] [PubMed]
- Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods 2010;50:244-9. [Crossref] [PubMed]
- Guo C, Sah JF, Beard L, et al. The noncoding RNA, miR‐126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3‐kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008;47:939-46. [Crossref] [PubMed]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101-8. [Crossref] [PubMed]
- Xiao B, Guo J, Miao Y, et al. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta 2009;400:97-102. [Crossref] [PubMed]
- Liu H, Zhu L, Liu B, et al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett 2012;316:196-203. [Crossref] [PubMed]
- Cook NR. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin Chem 2008;54:17-23. [Crossref] [PubMed]
- Wang W, Li F, Zhang Y, et al. Reduced expression of miR-22 in gastric cancer is related to clinicopathologic characteristics or patient prognosis. Diagn Pathol 2013;8:102. [Crossref] [PubMed]
- Tsujiura M, Ichikawa D, Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 2010;102:1174-9. [Crossref] [PubMed]
- Yao Y, Suo AL, Li ZF, et al. MicroRNA profiling of human gastric cancer. Mol Med Rep 2009;2:963-70. [PubMed]
- Guo J, Miao Y, Xiao B, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 2009;24:652-7. [Crossref] [PubMed]
- Li X, Luo F, Li Q, et al. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol Rep 2011;26:1431-9. [PubMed]
- Konishi H, Ichikawa D, Komatsu S, et al. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer 2012;106:740-7. [Crossref] [PubMed]
- Rotkrua P, Akiyama Y, Hashimoto Y, et al. MiR-9 downregulates CDX2 expression in gastric cancer cells. Int J Cancer 2011;129:2611-20. [Crossref] [PubMed]


