Single nucleotide polymorphism rs4648298 in miRNAs hsa-miR21 and hsa-miR590 binding site of COX gene is a strong colorectal cancer determinant
Introduction
Colorectal cancer (CRC) rated as the third diagnosed malignancy and fourth most common cause of cancer mortality worldwide. In developed regions, the occurrence of CRC for men and women increases to 37.7 and 24.3 per 100,000 persons per year (1,2). However the incidence of CRC is lower in Asian than in western countries, recent studies have shown, increasing rates of CRC in Asia and especially in Iran as a developing country (3,4). Although previous reports revealed that many environmental and lifestyle factors such as high body mass index (BMI), smoking, low activity and alcohol drinking increase the risk of CRC (5,6). Albeit, from all people who are exposed to same risk factors just some of them developed CRC. It could interpret that CRC is a complex disease, influenced by genetics and environmental factors. MicroRNAs constitute a novel discovered class of small non-coding RNAs (about 22 nucleotides) that play important roles in the regulation of gene expression by binding to the 3'-untranslated regions (3'-UTR) of specific mRNAs, and leading to mRNA cleavage or translational repression. Accumulated evidences demonstrated that miRNAs are one of the key players in cell differentiation, growth, apoptosis and insulin secretion (7,8). The most frequent type of variations in the human genome is single nucleotide polymorphism (SNP) that they exist once almost in every 300 nucleotides. Several studies demonstrated that SNPs in 3'UTR of mRNA may impact microRNAs functions by influence in secondary structure of 3'UTR and thermodynamic features of hybridization site (7,9,10). These SNPs can deregulate expression of target gene by change in binding capacity of miRNAs. The association between inflammation and tumorigenesis especially in CRC is well-established. COXs are rate-limiting enzymes that mediate the formation of prostaglandins from arachidonic acid (11,12). Two major cyclooxygenase isoforms are Cyclooxygenase-1 (COX-1) and COX-2 with 60% homology, most tissues express COX-1 at relatively low levels (13). Cyclooxygenase-2 (COX-2) is the inducible isoform that is not found in most normal tissues but can be induced by cytokines (inflammatory reactions), growth factors or tumor promoters (14-17). In overall COX-2 could be known as an acute response protein that is expressed as part of respond to inflammation (18). Overexpression of COX-2 is sufficient for carcinogenesis in animal models and blockage of the COX-2 pathway results in decrease in tumor occurrence and progression (19). COX-2 overexpression is observed in multiple cancers especially sporadic CRC (20,21). Several studies indicate that SNPs in the COX-2 gene may modulate the risk of CRC development (12,14,22). As mentioned polymorphisms in 3'UTR of COX-2 gene may affect the expression by modifying of miRNAs binding capacity. In this case control study we assessed association between SNP rs4648298 in 3'UTR of COX2 gene with sporadic CRC risk in Iranian population. Investigate on miRNA and SNP databases (such as miRBase, miRanda and mirdsnp) shows that this polymorphism (rs4648298) is in miRNA binding site of hsa-miR21, hsa-miR590 and in vicinity of has-miR101 binding site that could influence on miRNA-mRNA interaction. Non-steroidal anti-inflammatory drugs (NSAIDs) known as reducer of CRC risk due to their significant roles in decreasing of inflammation, especially by inhibition of COX-2 expression; therefore in this study we also assessed interaction between SNP rs4648298 and use of NSAIDs in our case—control study. Moreover, we compared our results with previously published results for rs4648298 using meta-analysis.
Methods
Study population
For this case control study, during the 2-year period from mid-2013 to mid-2015, totally 176 participants were selected amongst patients referred to the colonoscopy centers of the University Hospitals, Isfahan, Iran. CRC cases, 88, were first diagnosed by colonoscopy and then followed their pathology report for the ultimate confirmation of their colonoscopy based CRC diagnosis. Controls, 88, were also selected from the same population with no signs of CRC detected in colonoscopy examination. The study was approved by Isfahan University of Medical Sciences Ethics board (approval number 392263) and written informed consent was filled and signed by all participants. To eliminate any established genetic risk factor for CRC, patients were individually interviewed to select cases with sporadic CRC without positive history of familial cancers. Also all participants were asked to fill up a questionnaire in order to register the parameters known to influence the CRC susceptibility risk including gender, age, BMI, physical activity, smoking status and NSAIDs consumption.
Genotyping of polymorphism
Approximately, 5 mL venous blood was collected from each participant. DNA was extracted by Prime Prep Genomic DNA Isolation Kit (GeNet Bio). The quality and quantity of the extracted DNA was assessed by agarose gel electrophoresis and spectroscopy, and then DNA was stored at −20 °C until analyzed. Polymorphism of COX-2 gene was genotyped by PCR-RFLP method. Using appropriate primer set, 392 bp fragment of COX-2 gene that contained the selected polymorphism was amplified by PCR. Thermal cycling contained an initial denaturation of 94 °C for 4 min and then 33 cycles including 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 30 s after these cycles final extension of 72 °C for 5 min. Restriction fragment length polymorphism (RFLP) was carried out using the AluI restriction endonuclease. PCR product size, restriction enzyme used, fragments subsequent to digestion and primer sequences used, are given in the Table 1. Finally the result of genotyping was confirmed by randomly selected samples for direct sequencing.

Full table
Meta-analysis
Methods and search strategy
We had intended to summarize data on relationship between SNP rs4648298 in 3'UTR of COX-2 gene and cancer risk.
In this field, we conducted a literature search, up to Jul 2017, of MEDLINE, Cochrane review, Google Scholar and Scopus databases for related studies. The search terms were “Neoplasms” (MeSH) AND “SNPs” (MeSH) AND “rs4648298” (tiab) AND “COX-2” (tiab).
Studies were eligible for inclusion in the current analysis if: (I) SNP rs4648298 in 3'UTR of COX-2 gene was assessed; (II) their final outcome was cancer risk; and (III) estimates of relative risks (RR), hazard ratios (HR) or odds ratio (OR) with corresponding 95% confidence interval (CI) for the cancer risk among SNP genotypes were provided. Two investigators extracted data independently, and any discrepancies were resolved by discussion.
Statistical analysis
Odds ratio were used as the measure association between SNP rs4648298 in 3'UTR of COX-2 gene and cancer risk. Meta-analyses were performed using the random-effects model that was presented as forest plots with 95% CI. The statistical heterogeneity of the studies was calculated with the Cochran’s Q and I2 index (I2>50% was considered as significant heterogeneity). To assess the impact of possible factors on pooled effect size and heterogeneity, subgroup analyses were performed. Publication bias was assessed using the funnel plot method and Egger’s test. Statistical analyses were conducted using the statistical software package Stata version 11.2. P values less than 0.05 were considered statistically significant.
Statistics
Genotype frequencies in case and controls were tested for Hardy-Weinberg equilibrium using χ2 test. Associations between COX-2 (rs4648298) polymorphism with susceptibility to sporadic CRC were examined by logistic regression analysis. Correlation among this polymorphism and CRC was evaluated using ORs and 95% CIs. The significance level was set at P<0.05. The difference in demographic and lifestyle characteristics distribution such as age, gender, smoking status, BMI and NSAIDs consumption, assessed by Pearson χ2 test for categorical variables and t-test for continuous variables. Mann-Whitney test were used to compare physical activity between CRC and control groups. All statistical analyses were performed using SPSS (SPSS, Chicago, IL, USA).
Results
Demographic and lifestyle characteristics
To evaluate correlation between risk factors (genetic and environmental) with CRC incidence, we analyzed 176 total participants in case and control groups; 88 patients (40 males and 48 females) in case and 88 (45 males and 43 females) healthy subjects in control group. There was no significant association between case and control group regarding age (P=0.56) and gender (P=0.45), this illustrating that for these variables matching was adequate. Also for smoking status (P=0.50) and mean BMI (P=0.33) we could not found any difference among two groups. However we found significant difference between control and case group in term of physical activity (OR =0.58, 95% CI, 0.41–0.83). Also this study demonstrated that individuals in control group were more NSAIDs consumer compared to patient group (OR =0.30, 95% CI, 0.13–0.68). Characteristics of study population are summarized in Table 2.
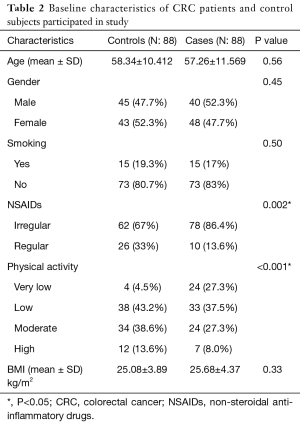
Full table
Genotype and allele distribution
The genotype distribution of rs4648298 polymorphism in two groups was in Hardy-Weinberg equilibrium. Genotype distribution in current study indicated that homozygous variant (GG) was not found in case and control group. The frequency of AA genotype in case and control group was 64.8% and 93.2% respectively and frequency of AG genotype in case and control group was 35.2% and 6.8% respectively (Table 3). In this evaluation we found significant association between AA genotype and decrease risk of CRC (OR =0.14; 95% CI, 0.05–0.34; P<0.001). Also in allele distribution we found that A allele have high frequency in control group compared to case group and we found that A allele was associated with decrease risk of CRC (OR= 0.17; 95% CI, 0.07–0.04; P<0.001). Furthermore, we assessed association between COX-2 polymorphism, rs4648298, genotypes and sporadic CRC risk in subgroups of participants that categorized by age (<55 and ≥55), gender (male and female), smoking status (smoker and nonsmoker) and NSAIDs consuming (regular and irregular) (Table 4). Data indicated that there was significant difference in each sub group of age, NSAIDs consuming and gender between case and control group regarding to CRC risk. This means that in these subgroups, AA genotype have protective effect for CRC similar to overall. However in smoker subgroup there was no association between this polymorphism and CRC risk (P=0.17), but in nonsmoker subgroup AA genotype was correlated with decrease risk of CRC (P<0.001).

Full table
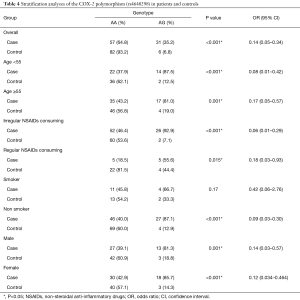
Full table
Findings from meta-analysis
We identified nine studies including gastric adenocarcinoma (23), CRC (14,15,24-26), breast cancer and prostate cancer (27) and lymphoma (28) that met the inclusion criteria of this meta-analysis. The cancer risk for AG + GG and AG genotypes compared to AA SNPs was assessed.
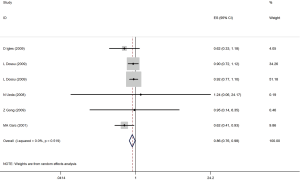
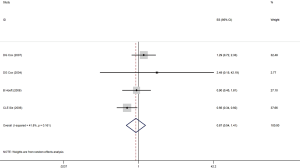
In a meta-analysis on six risk estimates for the AG versus AA genotype, we found a significant inverse association between AG SNPs and risk of gastric adenocarcinoma, CRC, breast cancer and prostate cancer (OR =0.86; 95% CI, 0.76–0.98; P<0.02; Figure 1). Publication bias was not observed by Egger’s test (P=0.49). Also, there were not asymmetries in funnel’s plots in the current analyses. Also, a between-study heterogeneity was not found (I2=0.0%, P=0.519).
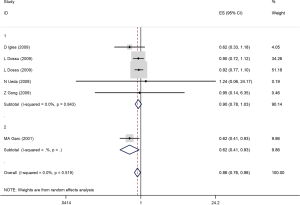
Through subgroup analysis by studies design, among case-control studies (ID =1), we found that a non-significant protective effects in AG compared to AA polymorphism in cancers risk (OR =0.90; 95% CI, 0.87–1.03; P=0.11). However, there was a significant association between AG genotype and gastric adenocarcinoma in the MA Garc study with cohort design (OR =0.62; 95% CI, 0.41–0.93; P=0.02; Figure 2). Furthermore, individuals with AG + GG variant in rs4648298 in 3'UTR of COX-2 gene compared with AA had not a significant less odd for CRC, breast cancer and lymphoma among four studies (OR =0.87; 95% CI, 0.54–1.41; P<0.57; Figure 3). The test of between-studies heterogeneity was not significant (I2=41.8%, P=0.161). Also, funnel plots and Egger’s test (P=0.40) indicated no publication bias in studies.
Discussion
To the best of our knowledge present study is the first report in Iranian population that investigates the correlation between COX-2 polymorphism, rs4648298, with CRC risk. COX-2 or PTGS2 enzyme as an inducible isoform of cyclooxygenase is involved in prostaglandin synthesis from arachidonic acid. Induction of COX-2 by growth factors, inflammatory cytokines and tumor promoters lead to production of prostaglandins that could stimulate the inflammation, cell proliferation, angiogenesis and differentiation (29,30). Previous study demonstrated that COX-2 is overexpressed in both sporadic colorectal adenomas and carcinomas (31). On the other hand a growing body of evidence proposed that NSAIDs are associated with decrease risk of CRC by inhibition of COX-2 (32-34).
MicroRNAs (miRNAs) are small non-coding RNAs involved in posttranslational gene expression regulation. Base pairing with specific sequence at 3'UTR of target mRNAs is key feature for miRNAs to execute this regulatory function (7,8). SNPs located in miRNA binding site contribute to the disruption of miRNA recognition elements (MREs) or create new sites resulting in up- or down-regulation of target genes (35,36). In addition previous studies revealed that in 3'UTR of COX-2 gene there were AU-rich elements which are binding sites for proteins and inflammatory mediators that could modulate stability of COX-2 mRNA thus SNPs in this region could disturb these interactions (37,38).
In current study we evaluate association between a COX-2 polymorphism (rs4648298) and CRC risk in Iranian population. This polymorphism is located in the recognition position of miR-590, miR-21 and vicinity of miR-101 binding sites at 3'UTR of COX-2 gene thus could affect on binding capacity of this miRNAs. Our results demonstrated that AA genotype and A allele were more frequent in control group compared with AG genotype and G allele. It could be interpreted that AA genotype and A allele have protective effect for healthy subjects (PAA and A: <0.001).
Also the results obtained from the meta-analysis showed a significant inverse association between AG SNPs and risk of all cancers. Several studies carried out on association between this polymorphism and risk of different cancer types. Two reports in Brazilian and Caucasian populations demonstrated that there was no association between this polymorphism and breast cancer risk (39,40), also in a study in North Carolina (USA) on colorectal adenoma no association was found (15). In Spanish population G allele was reported to be associated with longer survival and AG genotype was associated with clinicopathological features and rs4648298 (AG genotype) known as a good prognostic indicator for CRC patients (14). While Cox et al. in same population indicated that individuals with AG + GG genotypes have higher risk of CRC (24). Also in Caucasians AA genotype was correlated with advance stage and decrease of overall survival in patient with gastric adenocarcinoma (23). Therefore based on our results protective effect for AA genotype and A allele in healthy subjects was not in agreement with previous studies. The reasons for the contradictory results may be due to differences in ethnic background and different types of cancers. However, performing replicative studies in every population is necessary to validate these results. Stratification of participants based on age, sex, smoking status and especially by NSAIDs consuming revealed that in each subgroup AA genotypes and A allele have protective effects for control subjects except in smoker subgroup. However previous studies reported, there were significant interactions between NSAIDs consuming and COX-2 variant. Some of these gene polymorphisms could modify the effect of NSAIDs in the prevention of CRC (11,24). In our study the same genotype in regular and irregular subgroups has been associated with CRC risk, therefore it could be interpret that this polymorphism did not effect on NSAIDs response. In this study may be there were some possible limitations in statistical validity of our results such as small sample size, so further association studies in a large sample size would be helped to confirm suggested correlations. Moreover other variants that were not included in this study may be involved in determining the risk of CRC and NSAIDs interaction, thus future studies are necessary.
Acknowledgements
Financial support of Acquired Immunodeficiency Research Center, Isfahan University of Medical Sciences, Isfahan, Iran is gratefully acknowledged. We also appreciate cooperation of all voluntaries participated in this study.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by Isfahan University of Medical Sciences Ethics board (approval number 392263) and written informed consent was filled and signed by all participants.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Aykan NF, Yalçın S, Turhal NS, et al. Epidemiology of colorectal cancer in Turkey: A cross-sectional disease registry study (A Turkish Oncology Group trial). Turk J Gastroenterol 2015;26:145-53. [Crossref] [PubMed]
- Kolahdoozan S, Sadjadi A, Radmard AR, et al. Five common cancers in Iran. Arch Iran Med 2010;13:143-6. [PubMed]
- Hosseini SV, Izadpanah A, Yarmohammadi H. Epidemiological changes in colorectal cancer in Shiraz, Iran: 1980-2000. ANZ J Surg 2004;74:547-9. [Crossref] [PubMed]
- Zapatier J, Avalos D, Tandon K, et al. Can adjusting BMI for age and sex provide for a better predictor of colonic neoplasia? Eur J Gastroenterol Hepatol 2015;27:974-80. [Crossref] [PubMed]
- Poomphakwaen K, Promthet S, Suwanrungruang K, et al. Risk factors for colorectal cancer in Thailand. Asian Pac J Cancer Prev 2015;16:6105-9. [Crossref] [PubMed]
- Mosallayi M, Simonian M, Khosravi S, et al. Polymorphism (rs16917496) at the miR-502 Binding Site of the Lysine Methyltransferase 5A (SET8) and Its Correlation with Colorectal Cancer in Iranians. Adv Biomed Res 2017;6:77. [PubMed]
- Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 2009;7:147-54. [Crossref] [PubMed]
- Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 2008;29:579-84. [Crossref] [PubMed]
- Bhaumik P, Gopalakrishnan C, Kamaraj B, et al. Single nucleotide polymorphisms in microRNA binding sites: implications in colorectal cancer. ScientificWorldJournal 2014;2014:547154. [PubMed]
- Daraei A, Salehi R, Mohamadhashem F. PTGS2 (COX2) -765G>C gene polymorphism and risk of sporadic colorectal cancer in Iranian population. Mol Biol Rep 2012;39:5219-24. [Crossref] [PubMed]
- Pereira C, Medeiros RM, Dinis-Ribeiro MJ. Cyclooxygenase polymorphisms in gastric and colorectal carcinogenesis: are conclusive results available? Eur J Gastroenterol Hepatol 2009;21:76-91. [Crossref] [PubMed]
- Garavito RM, Malkowski MG, DeWitt DL. The structures of prostaglandin endoperoxide H synthases-1 and -2. Prostaglandins Other Lipid Mediat 2002;68-69:129-52. [Crossref] [PubMed]
- Iglesias D, Nejda N, Azcoita MM, et al. Effect of COX2 -765G>C and c.3618A>G polymorphisms on the risk and survival of sporadic colorectal cancer. Cancer Causes Control 2009;20:1421-9. [Crossref] [PubMed]
- Gong Z, Bostick RM, Xie D, et al. Genetic polymorphisms in the cyclooxygenase-1 and cyclooxygenase-2 genes and risk of colorectal adenoma. Int J Colorectal Dis 2009;24:647-54. [Crossref] [PubMed]
- Prescott SM, Fitzpatrick FA. Cyclooxygenase-2 and carcinogenesis. Biochim Biophys Acta 2000;1470:M69-78. [PubMed]
- Cao Y, Prescott SM. Many actions of cyclooxygenase-2 in cellular dynamics and in cancer. J Cell Physiol 2002;190:279-86. [Crossref] [PubMed]
- Martín Sanz P, Hortelano S, Bosca L, et al. Cyclooxygenase 2: understanding the pathophysiological role through genetically altered mouse models. Front Biosci 2006;11:2876-88. [Crossref] [PubMed]
- Williams CS, Tsujii M, Reese J, et al. Host cyclooxygenase-2 modulates carcinoma growth. J Clin Invest 2000;105:1589-94. [Crossref] [PubMed]
- Fujimura T, Ohta T, Oyama K, et al. Role of cyclooxygenase-2 in the carcinogenesis of gastrointestinal tract cancers: a review and report of personal experience. World J Gastroenterol 2006;12:1336-45. [Crossref] [PubMed]
- van Rees BP, Ristimäki A. Cyclooxygenase-2 in carcinogenesis of the gastrointestinal tract. Scand J Gastroenterol 2001;36:897-903. [Crossref] [PubMed]
- Ashktorab H, Tsang S, Luke B, et al. Protective effect of Cox-2 allelic variants on risk of colorectal adenoma development in African Americans. Anticancer Res 2008;28:3119-23. [PubMed]
- García-González MA, Nicolás-Pérez D, Lanas A, et al. Prognostic role of host cyclooxygenase and cytokine genotypes in a Caucasian cohort of patients with gastric adenocarcinoma. PloS one 2012;7:e46179. [Crossref] [PubMed]
- Cox DG, Pontes C, Guino E, et al. Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br J Cancer 2004;91:339-43. [Crossref] [PubMed]
- Siezen CL, Bueno-de-Mesquita HB, Peeters PH, et al. Polymorphisms in the genes involved in the arachidonic acid-pathway, fish consumption and the risk of colorectal cancer. Int J Cancer 2006;119:297-303. [Crossref] [PubMed]
- Ueda N, Maehara Y, Tajima O, et al. Genetic polymorphisms of cyclooxygenase-2 and colorectal adenoma risk: the Self Defense Forces Health Study. Cancer Sci 2008;99:576-81. [Crossref] [PubMed]
- Dossus L, Kaaks R, Canzian F, et al. PTGS2 and IL6 genetic variation and risk of breast and prostate cancer: results from the Breast and Prostate Cancer Cohort Consortium (BPC3). Carcinogenesis 2010;31:455-61. [Crossref] [PubMed]
- Hoeft B, Becker N, Deeg E, et al. Joint effect between regular use of non-steroidal anti-inflammatory drugs, variants in inflammatory genes and risk of lymphoma. Cancer Causes Control 2008;19:163-73. [Crossref] [PubMed]
- Williams CS, Mann M, DuBois RN. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 1999;18:7908-16. [Crossref] [PubMed]
- Patrignani P, Tacconelli S, Sciulli MG, et al. New insights into COX-2 biology and inhibition. Brain Res Brain Res Rev. 2005;48:352-9. [Crossref] [PubMed]
- Eberhart CE, Coffey RJ, Radhika A, et al. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology 1994;107:1183-8. [Crossref] [PubMed]
- Bosetti C, Gallus S, La Vecchia C. Aspirin and cancer risk: an updated quantitative review to 2005. Cancer Causes Control 2006;17:871-88. [Crossref] [PubMed]
- Koehne CH, Dubois RN. COX-2 inhibition and colorectal cancer. In: Seminars in oncology; Amsterdam: Elsevier 2004;31:12-21.
- Wang D, DuBois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene 2010;29:781-8. [Crossref] [PubMed]
- Ahangari F, Salehi R, Salehi M, et al. A miRNA-binding site single nucleotide polymorphism in the 3'-UTR region of the NOD2 gene is associated with colorectal cancer. Med Oncol 2014;31:173. [Crossref] [PubMed]
- Yu Z, Li Z, Jolicoeur N, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res 2007;35:4535-41. [Crossref] [PubMed]
- Jing Q, Huang S, Guth S, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005;120:623-34. [Crossref] [PubMed]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 2006;33:7138-50. [Crossref] [PubMed]
- Cox DG, Buring J, Hankinson SE, et al. A polymorphism in the 3'untranslated region of the gene encoding prostaglandin endoperoxide synthase 2 is not associated with an increase in breast cancer risk: a nested case-control study. Breast Cancer Res 2007;9:R3. [Crossref] [PubMed]
- Piranda DN, Festa-Vasconcellos JS, Amaral LM, et al. Polymorphisms in regulatory regions of cyclooxygenase-2 gene and breast cancer risk in Brazilians: a case-control study. BMC Cancer 2010;10:613. [Crossref] [PubMed]


