Immune profiling of microsatellite instability-high and polymerase ε (POLE)-mutated metastatic colorectal tumors identifies predictors of response to anti-PD-1 therapy
Introduction
Early evidence supporting the concept of programmed cell death 1 (PD-1) and programmed death ligand 1 (PD-L1) blockade as a form of cancer immunotherapy arose from preclinical studies demonstrating that activation of the PD-1/PD-L1 axis suppressed the activation and proliferation of tumor antigen-specific T cells and promoted tumorigenesis, which was reversed with PD-1/PD-L1 blockade (1,2). Initial phase I studies investigating several humanized monoclonal IgG4 antibodies targeting PD-1 and PD-L1 in advanced solid tumors were soon conducted and paved way for the development of the first PD-1 inhibitors, nivolumab and pembrolizumab, approved by the Food and Drug Administration (FDA) (3-5). In the studies that have followed, PD-L1 expression [most often ≥1% or ≥5% by immunohistochemistry (IHC)] was among the first candidate predictors of response to PD-1 blockade and has been associated with a 20–50% response rate to PD-1 inhibitors in select solid tumors (5-7). However, the documentation of PD-L1-negative patients with a response to anti-PD-1 therapy argues against the use of PD-L1 expression as the sole biomarker for selection (6).
Other studies have focused on the immune-infiltrating cells within the tumor microenvironment (TME) and have shown that pretreatment specimens from responders to PD-1/PD-L1 blockade have significantly higher densities of CD8+, PD-1+, and PD-L1+ T-cells at the invasive tumor margin and tumor parenchyma (with close proximity between PD-1 and PD-L1 expression) compared to nonresponders (8,9). Furthermore, the CD8+ tumor-infiltrating lymphocytes (TILs) in responders have a more clonal T-cell antigen receptor (TCR) repertoire, high expression of cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and PD-1 (PD-1hiCTLA-4hi) consistent with a partially exhausted CD8+ T-cell phenotype capable of producing IFNγ, high expression of major histocompatibility complex (MHC) class II antigen HLA-DR, reduction in the CD4+/CD8+-cell ratio, and a gene expression profile consistent with activation of CD8+ and Th1 T-cell responses (8-13).
Recently, next-generation sequencing (NGS) has uncovered that high mutational load can also predict benefit from immunotherapy due to the immunogenic nature of neoantigens generated from an increased burden of somatic mutations (14-16). In addition, microsatellite instability (MSI) or mismatch repair (MMR) deficiency has been shown to predict clinical benefit from immune checkpoint blockade (17). Notably, genomic analysis confirmed a significantly higher mutational load in MMR-deficient tumors than MMR-proficient tumors that was associated with prolonged progression-free survival (PFS). These findings have been corroborated in other studies demonstrating that MSI with high tumor mutational burden (TMB) due to defective MMR can predict response to anti-PD-1, anti-PD-L1, and anti-CTLA-4 therapy in metastatic colorectal cancer (mCRC) (18-21).
Beyond the hypermutated phenotype of MSI-high (MSI-H) tumors, germline and somatic mutations in the DNA proofreading enzyme polymerase ε (POLE) contribute to an ultramutated tumor phenotype due to an extremely high rate of base substitution mutations (22,23). POLE-mutated tumors comprise a phenotype mutually exclusive of MSI-H and microsatellite stable (MSS) tumors with the highest mutational burden among the 3 phenotypes (24-27). Similar to MSI-H tumors, POLE-mutated tumors are highly immunogenic due to enrichment by mutation-associated neoantigens and have demonstrated response to checkpoint inhibitors in non-small cell lung cancer (NSCLC) and endometrial cancer (15,26-30). Despite the clearly increased immunogenicity of MSI-H and POLE-mutated colorectal cancer, no studies to our knowledge have investigated this combined set of patients for predictive markers of response to checkpoint inhibition.
We previously conducted an analysis of tumors from mCRC patients at our institution that underwent comprehensive genomic profiling using NGS and identified a subset of patients with MSI-H and POLE-mutated tumors that could potentially benefit from PD-1 blockade (31). We subsequently reported an initial case of a treatment-refractory and MSS mCRC harboring a POLE mutation with excellent response to PD-1 blockade (32). We now describe our single-institution experience involving a cohort of patients with MSI-H and POLE-mutated metastatic colorectal tumors treated with checkpoint inhibitors and report on candidate predictive markers of response to PD-1 blockade.
Methods
Study patients and tumor samples
Patients diagnosed with advanced or metastatic (stage IV) colorectal cancer treated at the Gastrointestinal Medical Oncology Clinic at City of Hope National Medical Center (Duarte, CA, USA) between April 2013 and January 2017 were screened for eligibility. Eligible patients must have had pathologically confirmed advanced or metastatic colon or rectal cancer, documented MSI-H (by previously defined methods) or POLE-mutated tumors (by comprehensive tumor genomic profiling via FoundationOne, Foundation Medicine, Inc., Cambridge, MA, USA), and treatment with an anti-PD-1 or an anti-PD-L1, with or without an anti-CTLA-4 agent for progressive metastatic disease (33). There were no exclusions to previous treatment or lines of prior therapy, tumor histology, medical comorbidities, or performance status. The study was approved by the City of Hope Institutional Review Board (IRB) under protocol # I16406, “Exploratory Tumor Immuno-profiling of Microsatellite Instability and POLE Hyper-mutated Colorectal Cancer.” This study is consent exempt as per the IRB and therefore no patient informed consent was obtained.
Study design
Retrospective analysis of genomic alterations in our cohort of mCRC patients was performed through reports provided by Foundation ICE (Interactive Cancer Explorer). Patient characteristics were obtained by chart abstraction. Response to anti-PD-1, anti-PD-L1, or anti-CTLA-4 therapy was defined according to Response Evaluation Criteria in Solid Tumors (RECIST) criteria and obtained from medical records and confirmed by the investigators (34). Patients were categorized as responders to checkpoint inhibition if the best overall response was a partial response (PR) or complete response (CR) and nonresponders if the best overall response was stable disease (SD) or progressive disease (PD) from the start of immunotherapy to the cut-off date of May 4, 2017.
Multispectral fluorescent IHC and image analysis
Formalin-fixed paraffin-embedded (FFPE) specimens were cut into 3 µm sections and mounted on glass slides. For each specimen, 4 multispectral IHC panels were stained using PerkinElmer Opal kit. All slides were scanned using the Vectra Automated Quantitative Pathology Imaging System which uses excitation and emission spectra for each individual fluorophore and 200× magnification images were taken and analyzed using the image analysis software inForm (PerkinElmer, Hopkinton, MA, USA) (35). Panel 1 included CD8 (SP16, Biocare), PD-1 (NAT105, Biocare), PD-L1 (SP142, SpringBio) cytokeratin 20 (CK20, Ks20.8, Dako), and 4',6-diamidino-2-phenylindole (DAPI, PerkinElmer); panel 2 included CD68 (kp1, Biocare), PD-L1, CK20, and DAPI; panel 3 included CD4 (4B12, Agilent), FOXP3 (236A/E7, Biocare), and DAPI; panel 4 included CD4, T-bet (EPR9301, Abcam), and DAPI.
Statistical analyses
The sample size was determined by the total number of mCRC patients with confirmed MSI-H or POLE-mutated tumors. Differences between tumor-infiltrating immune cell densities were compared using unpaired T test performed in Graphpad Prism (V7.0) and P values of less than 0.05 were considered statistically significant.
Results
Study population
From April 2013 to January 2017, a total of 237 mCRC patients treated at our single institution who had undergone comprehensive genomic profiling of their tumors by NGS (FoundationOne®) were screened. A cohort of 8 patients (5 with MSI-H tumors and 3 with POLE-mutated tumors) with sufficient archival tissue for correlative studies and who received anti-PD-1, anti-PD-L1, or anti-CTLA-4 therapy were identified (Table 1).
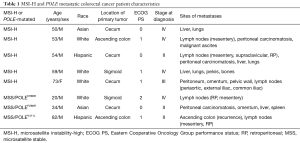
Full table
Treatment characteristics and response
Individual treatment details and treatment outcomes are detailed in Table 2. Seven patients received pembrolizumab (anti-PD-1 monoclonal antibody) monotherapy at a fixed dose of 200 mg intravenously every 3 weeks (87.5%). One patient with an MSI-H tumor received investigational therapy targeting the PD-1/CTLA-4 pathway. Four out of 8 patients (50%) were responders to checkpoint inhibition (PR or CR as the best overall response) and 50% were nonresponders (1 SD and 3 PD as the best overall response). The majority of patients were heavily pretreated with immune profiling conducted on archival tissues obtained from the primary tumor in all patients except for 1. The time from archival tissue collection to initiation of checkpoint inhibitor therapy ranged from 2 months to 5 years (Table 2).
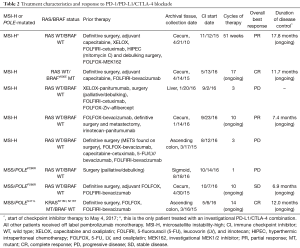
Full table
Immune profiling
Tumor specimens from all 8 mCRC patients (5 MSI-H and 3 POLE-mutated) were subjected to multispectral immunofluorescent IHC staining and image analysis. Among the 4 responders with MSI-H and POLE-mutated metastatic colorectal tumors, a large amount of CD8+ T-cells with >80–90% of these expressing PD-1 were identified (Figure 1). In the 4 nonresponders to PD-1 blockade (2 MSI-H and 2 POLE-mutated), the population of CD8+ T-cells and PD-1-expressing CD8+ TILs were relatively sparse compared to responders (Figure 1). In 3 out of 4 responders with MSI-H or POLE-mutated mCRC, the population of PD-1-expressing CD8+ T-cells primarily infiltrated the tumor stroma. One complete responder with a MSI-H tumor showed high PD-1+ CD8+ T-cell infiltration both inside the tumor parenchyma and stroma (Figure 1). The density of PD-1+ CD8+ TILs was significantly higher in pretreatment specimens from responders compared to nonresponders (P=0.0007, Figure 2).
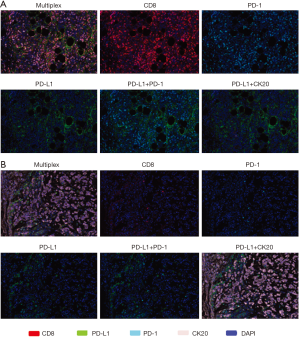
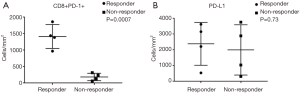
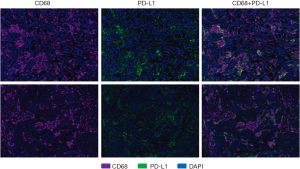
PD-L1 expression was more varied across responders and nonresponders. When observed, PD-L1 expression occurred in close proximity to PD-1 expression (Figure 1). PD-L1 expression was primarily observed on the immune-infiltrating cells within the TME with a small portion of PD-L1 expression observed on tumor cells. Using a multi-marker panel including CD68, PD-L1, CK20, and DAPI, subsequent image analysis demonstrated that non-tumor PD-L1 expression was observed in CD68+ tumor-associated macrophages (Figure 3). In the 4 responders to checkpoint inhibitors, 3 patients (2 MSI-H and 1 POLE-mutated) showed PD-L1 expression on CD68+ tumor-associated macrophages only, while 1 MSI-H patient showed PD-L1 expression on both tumor cells and tumor-associated macrophages, with less than 5% of PD-L1 expressed on tumor cells. In the 4 nonresponders to PD-1 blockade (2 MSI-H and 2 POLE-mutated), PD-L1 was found to be expressed on CD68+ tumor-associated macrophages (Figure 3). Ultimately, levels of PD-L1 expression were not significantly different between responders and nonresponders with MSI-H or POLE-mutated metastatic colorectal tumors (P=0.73, Figure 2).
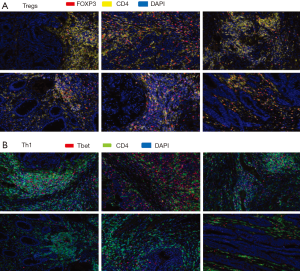
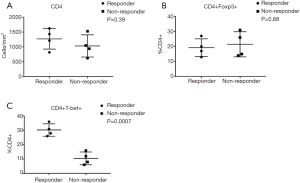
To further elucidate the immune response characteristics between responders and nonresponders to PD-1 blockade in MSI-H and POLE-mutated metastatic colorectal tumors, a staining panel consisting of CD4, FOXP3 [marker of T-regulatory cells (Tregs)], and Tbet (marker of T-lymphocytes committed to the Th1 response) was used. Expression of CD4 and FOXP3 was observed within the TME of tumor specimens from both responders and nonresponders (Figure 4). However, densities of CD4+ T-cells (P=0.39) and the percentage of CD4+ FOXP3+ cells (P=0.68) were not significantly different between responders and nonresponders to PD-1/PD-L1/CTLA-4 blockade (Figure 5). A greater amount of CD4+ Tbet+ cells was identified within the TME of responders compared to nonresponders (Figure 4). Pretreatment tumor specimens of responders to checkpoint blockade showed a significantly higher percentage CD4+ Tbet+ T-cells infiltrating the TME when compared to those of nonresponders (P=0.0007, Figure 5).
Discussion
The purpose of this retrospective study was to perform immune profiling on pretreatment tumor specimens from a cohort of patients with metastatic colorectal tumors that are MSI-H or POLE-mutated and identify potential predictors of response to PD-1/PD-L1 blockade. We performed multispectral fluorescent IHC and image analysis on tumors from 8 mCRC patients (5 with MSI-H and 3 with POLE-mutated tumors) screened from a larger cohort of 237 mCRC patients who had undergone comprehensive genomic profiling by NGS at our institution. All 8 patients were treated with checkpoint inhibitors (7 with pembrolizumab and 1 with a combination of an anti-PD-L1 and anti-CTLA-4 agent).
We observed a large population of CD8+ T-cells within the TME of pretreatment specimens from responders in contrast to the relatively sparse amount of CD8+ T-cells in nonresponders (Figure 1). Furthermore, responders to checkpoint inhibitors had significantly higher densities of PD-1-expressing CD8+ TILs compared to nonresponders (P=0.0007). A recent study involving 40 resected MSI primary colorectal tumors identified that the TILs among an immunoreactive subset of samples frequently showed granzyme B and CD8 co-expression consistent with cytotoxic T-lymphocytes (CTLs) though CD8 and PD-1 co-expression was more variable (36). In a predominantly early-stage to locally-advanced CRC cohort, MSI tumors showed higher densities of TILs with CD8+ T-cells being most dramatically increased along with higher expression of PD-1, among other checkpoints, when compared to MSS tumors (29). High Immunoscore colorectal tumors, which are highly infiltrated by CD8+ T-cells at both the invasive margin and tumor center, have been shown to be significantly associated with MSI-H rather than MSS status, significantly overrepresented by cells expressing PD-1 at the tumor center and invasive margin, and significantly associated with improved survival in primarily localized CRC (37). Having a high Immunoscore at distant metastases appeared to correlate with improved survival in advanced CRC as well (38). Furthermore, responses to immunotherapy may be enhanced in MSI colorectal tumors based on recent evidence suggesting that frameshift mutations in ASTE1, HNF1A, and TCF7L2 genes have been associated with increased CD8+ TILs and can lead to production of immunogenic neoantigens recognized by specific CD8+ TILs (39). In predominantly non-metastatic colorectal and endometrial cancer patients, POLE-mutated tumors have similarly shown higher levels of CD8+ TILs and PD-1 expression than MSS tumors and are associated with a better oncological outcome when present in the setting of locoregional disease (24-27). However, data describing the relationship between CD8+ TILs/PD-1 expression and response to PD-1 blockade in MSI-H and POLE-mutated mCRC is relatively limited. Our findings are among the first to support that high tumor-infiltrating CD8+ T-cells and high PD-1-expressing CD8+ TILs can predict response to checkpoint inhibitors in this population. Our findings are consistent with other findings reported in immune-checkpoint responsive diseases, such as a melanoma (8-10,40). Further studies evaluating larger MSI-H/POLE cohorts are warranted to identify the threshold by which CD8+ TILs and PD-1+ co-expression predict response. Further understanding of the subsets of CD8+ TILs that are predictive of response, in addition to the overall population of tumor-infiltrating CD8+ T-cells, may enrich our selection of candidates for PD-1 blockade in MSI-H and POLE-mutated mCRC.
Notably, in 3 out of our 4 responders with MSI-H or POLE-mutated mCRC, the population of CD8+ TILs and CD8+ PD-1+ TILs predominantly infiltrated the tumor stroma though 1 responder with a MSI-H tumor showed high CD8+ and CD8+ PD-1+ T-cell infiltration both inside the tumor parenchyma and stroma. An increased population of CD8+ TILs has been observed at the invasive tumor front in responders to PD-1 blockade in MSI-H mCRC while high densities of CD8+ TILs have been found at the invasive tumor front and tumor stroma in predominantly non-metastatic MSI-H CRC (17,29). Further investigation is warranted in MSI-H and POLE-mutated CRC patients to assess whether the intensity of TILs across tumor compartments differs between responders and nonresponders and whether presence of TILs in both the tumor stroma and parenchyma correlate with prolonged responses to PD-1 blockade.
PD-L1 expression was more varied across responders and nonresponders in our MSI-H and POLE-mutated mCRC patients. When observed, PD-L1 expression occurred in close proximity to PD-1 expression. This is consistent with previous data describing a significant association between proximity of PD-1 and PD-L1 expression and response to PD-1 blockade (8). Furthermore, we observed that PD-L1 expression was found predominantly on the immune-infiltrating cells within the TME, specifically in CD68+ tumor-associated macrophages, while a minority of PD-L1 was expressed on tumor cells. Across several advanced malignancies, response to anti-PD-L1 therapy was more strongly associated with PD-L1 expression on tumor-infiltrating immune cells rather than on tumor cells (9). In CRC, the majority of cases of PD-L1 expression have been identified on immune-infiltrating cells as well (7). In MSI-H colorectal tumors, PD-L1 expression on tumor cells was virtually non-discernible while the majority of PD-L1 expression was found on myeloid cells (29,41). This is different from NSCLC, RCC, and melanoma specimens for which PD-L1 is more consistently expressed on tumor cells and infiltrating immune cells (7).
Mutations in POLE have been associated with higher expression of PD-L1 in endometrial cancer and CRC than MSS tumors, but correlation with response to PD-1 blockade has not been evaluated (24-27). Expression of PD-L1 has been associated with increased CD8+ TILs but may not be predictive of survival in early-stage MSI-H CRC; correlation of PD-L1 expression to anti-PD-1-therapy benefit has also not been studied in this population (42). We observed that there was no significant difference in levels of PD-L1 expression between responders and nonresponders (P=0.73) to checkpoint inhibitors in our MSI-H and POLE-mutated mCRC patients. Although classification of tumors based on PD-L1 expression and CD8+ T-cells has been proposed to inform selection for PD-1/PD-L1 inhibitors, CRC in general represents a tumor subtype with relatively lower amounts of CD8+ TILs and PD-L1 expression than NSCLC, RCC, and melanoma (43). Indeed, PD-L1 expression may not be as important in MSI-H mCRC as well based on a recent phase II trial that demonstrated response to PD-1 blockade in treatment-refractory MSI-H mCRC patients regardless of PD-L1 expression (44). Our findings provide support that PD-L1 expression may not predict response to checkpoint inhibitors in MSI-H and POLE-mutated mCRC patients though validation in larger, prospective studies is warranted. Further investigation may define other predictive biomarkers in this population. For example, mutations in JAK1/2 and transcriptional signatures have emerged as potential novel predictors of response to PD-1 blockade in melanoma (45,46).
A multi-marker panel consisting of CD4, FOXP3, and Tbet were used to further evaluate the immune profile in pretreatment specimens between responders and nonresponders to PD-1 blockade in our MSI-H and POLE-mutated mCRC patients. Densities of CD4+ T-cells within the TME were not significantly different between responders and nonresponders (P=0.39) in our cohort. This is concordant with findings from an advanced melanoma cohort that demonstrated that CD4 expression at baseline was not significantly associated with response to anti-PD-1 therapy (8). Additionally, the percentage of CD4+ FOXP3+ T-cells (marker of Tregs) within the TME did not significantly differ between responders and nonresponders in our study (P=0.68). In advanced melanoma, Tregs have not been implicated in predicting benefit to anti-PD-1 and anti-PD-L1 therapy (9,10). Treg-associated genes including FOXP3 in MSI-H colorectal tumors had similar levels of expression to those in MSS tumors (29). Our findings suggest that the number of CD4+ FOXP3+ T-cells present in the TME is not a predictor of response to anti-PD-1, anti-PD-L1, and anti-CTLA-4 therapy in metastatic colorectal tumors that are MSI-H or POLE-mutated.
In contrast, we observed that the percent of CD4+ Tbet+ T-cells (marker of Th1 response) in the TME was significantly higher in responders compared to nonresponders to checkpoint inhibition (P=0.0007). An expression pattern in tumors consistent with the generalized activation of CD8 and Th1 T-cell responses has been shown to significantly correlate with response to anti-PD-L1 therapy across several advanced malignancies (9). Notably, an activated Th1/CTL phenotype was identified in the TME of virtually all MSI-H colorectal tumors (29). Although increased expression of cytotoxic T-cell markers such as CD8A and other effector cytokines, when compared to MSS tumors, have been observed in predominantly early-stage colorectal and endometrial cancers harboring POLE mutations, their association with response to immunotherapy in POLE-mutated tumors is unknown (24-27). Our findings suggest that the presence of tumor-infiltrating immune cells with a Th1 phenotype within the TME may be predictive of response to checkpoint inhibitors in MSI-H and POLE-mutated mCRC patients. Our findings also reinforce the need to further identify T-cell subsets and phenotypes within the TME that could enrich our development of an ideal candidate biomarker for PD-1 blockade in MSI-H and POLE-mutated mCRC.
In conclusion, immune profiling of pretreatment tumor specimens in a cohort of MSI-H and POLE-mutated mCRC patients at our single-institution identified higher amounts of tumor-infiltrating CD8+ T-cells and PD-1 expressing CD8+ TILs in the TME of responders to PD-1 blockade when compared to nonresponders. Expression of PD-L1, CD4+ density, and Treg density within the TME are not predictive of response to PD-1 inhibitors. Significantly higher amounts of T-cells with a Th1 phenotype (CD4+ Tbet+ T-cells) are observed in the TME of responders compared to nonresponders. Further investigation of our findings in larger and, ideally, prospective settings are warranted to validate the candidacy of these potential biomarkers of response to checkpoint inhibitors in MSI-H and POLE-mutated metastatic colorectal tumors.
Acknowledgements
Funding: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572. PPL is supported by Stand Up to Cancer (SU2C). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the City of Hope Institutional Review Board (IRB) under protocol # I16406, “Exploratory Tumor Immuno-profiling of Microsatellite Instability and POLE Hyper-mutated Colorectal Cancer.” This study is consent exempt as per the IRB and therefore no patient informed consent was obtained.
References
- Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002;8:793-800. [Crossref] [PubMed]
- Iwai Y, Ishida M, Tanaka Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci USA 2002;99:12293-7. [Crossref] [PubMed]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [Crossref] [PubMed]
- Patnaik A, Kang SP, Tolcher AW, et al. Phase I study of MK-3475 (anti-PD-1 monoclonal antibody) in patients with advanced solid tumors. J Clin Oncol 2012;30:Abstr 2512.
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Sunshine J, Taube JM. PD-1/PD-L1 inhibitors. Curr Opin Pharmacol 2015;23:32-8. [Crossref] [PubMed]
- Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014;20:5064-74. [Crossref] [PubMed]
- Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71. [Crossref] [PubMed]
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Daud AI, Loo K, Pauli ML, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 2016;126:3447-52. [Crossref] [PubMed]
- Sznol M, Chen L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer. Clin Cancer Res 2013;19:1021-34. [Crossref] [PubMed]
- Taube JM, Anders RA, Young GD, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012;4:127ra37. [Crossref] [PubMed]
- Teng MW, Ngiow SF, Ribas A, et al. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res 2015;75:2139-45. [Crossref] [PubMed]
- Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;482:400-4. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Hochster HS, Bendell JC, Cleary JM, et al. Efficacy and safety of atezolizumab (atezo) and bevacizumab (bev) in a phase Ib study of microsatellite instability (MSI)-high metastatic colorectal cancer (mCRC). J Clin Oncol 2017;35:Abstr 673.
- Le DT, Uram JN, Wang H, et al. Programmed death-1 blockade in mismatch repair deficient colorectal cancer. J Clin Oncol 2016;34:Abstr 103.
- Lin EI, Tseng LH, Gocke CD, et al. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget 2015;6:42334-44. [Crossref] [PubMed]
- Overman MJ, Kopetz S, McDermott RS, et al. Nivolumab ± ipilimumab in treatment (tx) of patients (pts) with metastatic colorectal cancer (mCRC) with and without high microsatellite instability (MSI-H): CheckMate-142 interim results. J Clin Oncol 2016;34:Abstr 3501.
- Briggs S, Tomlinson I. Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 2013;230:148-53. [Crossref] [PubMed]
- Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 2013;45:136-44. [Crossref] [PubMed]
- Domingo E, Freeman-Mills L, Rayner E, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol 2016;1:207-16. [Crossref] [PubMed]
- Glaire MA, Domingo E, Vermeulen L, et al. POLE proofreading domain mutation defines a subset of immunogenic colorectal cancers with excellent prognosis. Ann Oncol 2016;27:149-206. [Crossref]
- Howitt BE, Shukla SA, Sholl LM, et al. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol 2015;1:1319-23. [Crossref] [PubMed]
- Mehnert JM, Panda A, Zhong H, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest 2016;126:2334-40. [Crossref] [PubMed]
- Bellone S, Centritto F, Black J, et al. Polymerase ε (POLE) ultra-mutated tumors induce robust tumor-specific CD4+ T cell responses in endometrial cancer patients. Gynecol Oncol 2015;138:11-7. [Crossref] [PubMed]
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43-51. [Crossref] [PubMed]
- van Gool IC, Eggink FA, Freeman-Mills L, et al. POLE proofreading mutations elicit an antitumor immune response in endometrial cancer. Clin Cancer Res 2015;21:3347-55. [Crossref] [PubMed]
- Gong J, Cho M, Sy M, et al. Molecular profiling of metastatic colorectal tumors using next-generation sequencing: A single-institution experience. Oncotarget 2017;8:42198-213. [Crossref] [PubMed]
- Gong J, Wang C, Lee PP, et al. Response to PD-1 blockade in microsatellite stable metastatic colorectal cancer harboring a POLE mutation. J Natl Compr Canc Netw 2017;15:142-7. [Crossref] [PubMed]
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248-57. [PubMed]
- Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205-16. [Crossref] [PubMed]
- Feng Z, Puri S, Moudgil T, et al. Multispectral imaging of formalin-fixed tissue predicts ability to generate tumor-infiltrating lymphocytes from melanoma. J Immunother Cancer 2015;3:47. [Crossref] [PubMed]
- Prall F, Hühns M. The PD-1 expressing immune phenotype of T cell exhaustion is prominent in the “immunoreactive” microenvironment of colorectal carcinoma. Histopathology 2017;71:366-74. [Crossref] [PubMed]
- Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show Immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698-711. [Crossref] [PubMed]
- Kwak Y, Koh J, Kim DW, et al. Immunoscore encompassing CD3+ and CD8+ T cell densities in distant metastasis is a robust prognostic marker for advanced colorectal cancer. Oncotarget 2016;7:81778-90. [Crossref] [PubMed]
- Maby P, Tougeron D, Hamieh M, et al. Correlation between density of CD8+ T-cell infiltrate in microsatellite unstable colorectal cancers and frameshift mutations: A rationale for personalized immunotherapy. Cancer Res 2015;75:3446-55. [Crossref] [PubMed]
- Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60-5. [Crossref] [PubMed]
- Kim JH, Park HE, Cho NY, et al. Characterisation of PD-L1-positive subsets of microsatellite-unstable colorectal cancers. Br J Cancer 2016;115:490-6. [Crossref] [PubMed]
- Rosenbaum MW, Bledsoe JR, Morales-Oyarvide V, et al. PD-L1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor-infiltrating lymphocytes. Mod Pathol 2016;29:1104-12. [Crossref] [PubMed]
- Ock CY, Keam B, Kim S, et al. Pan-Cancer immunogenomic perspective on the tumor microenvironment based on PD-L1 and CD8 T-cell infiltration. Clin Cancer Res 2016;22:2261-70. [Crossref] [PubMed]
- Overman MJ, Lonardi S, Leone F, et al. Nivolumab in patients with DNA mismatch repair deficient/microsatellite instability high metastatic colorectal cancer: update from CheckMate 142. J Clin Oncol 2017;35: abstr 519.
- Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35-44. [Crossref] [PubMed]
- Shin DS, Zaretsky JM, Escuin-Ordinas H, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 2017;7:188-201. [Crossref] [PubMed]


