An assessment of candidate genes to assist prognosis in gastric cancer
Introduction
Gastric cancer (GC) is the fourth commonest type of cancer and despite great improvements in treatment regimes, it still carries the second highest mortality rate for cancer worldwide (1,2). Over 90% of GC are adenocarcinomas, proximal (arising from the cardia) and distal (non-cardia tumours) with two main histological types: (I) well differentiated/ intestinal type, or (II) undifferentiated/diffuse type. Several aetiological factors in the development of GC have been identified, including Helicobacter pylori infection, high salt and low fruit and vegetable diets, smoking, obesity, radiation exposure, pernicious anaemia, blood group A and family history (1).
The presentation of GC is often delayed due to the non-specific nature of symptoms. Delayed presentation often results in more advanced disease at diagnosis, meaning curative surgery is not possible and palliative treatment becomes the treatment of choice (3,4). This unsurprisingly leads to an overall poor prognosis for GC. Five-year survival rates have been quoted ranging from 36–47% in large Western trials (5), with other sources quoting as low as 20% overall and 5% for metastatic disease (6).
Given these poor outcomes there is a drive for the earlier biomarker detection of GC, when the disease is at a more treatable stage. Carcinoembryonic antigen, carbohydrate antigen (CA) 19-9 and CA72-4 are perhaps the best-known tumour markers, although they are not ideal due to their relatively low sensitivity and specificity (20–30%) (7). Many genes have been identified as having potential roles in the determination of prognosis for GC and as potential or actual targets for chemotherapy.
Perhaps the best understood biomarkers for GC prognosis are E-cadherin and c-erbB2. E-Cadherin germline mutations were shown to result in depleted E-cadherin levels and the subsequent development of aggressive, poorly differentiated GC (8). c-erbB2 mutations meanwhile were shown to promote tumour invasion and metastasis, thus suggesting a potential for targeted therapy aimed at controlling disease spread (9).
A review by Kanda and Kodera has summarised recent research into potential GC biomarkers (7). They identified found multiple genes with altered expression or processing which have a role in early detection, recurrence (c-erbB2), predicting survival and predicting treatment response. Those overexpressed genes linked with predicting survival included cell surface proteins (B7-H4), adhesion factors (DPYSL3), transcription factors (MYCL1, YBX1), matrix proteins (SERPINA1) and cell cycle regulators (S100A6, CCND1). Down regulated genes which appeared to help predict survival included extracellular matrix proteins (ITIH5), tight junction assembly (JAMA), with SEMA3A and STUB1 implicated in tumour proliferation. They further identified genes with aberrant DNA methylation which can affect survival. These included phosphorylation inhibitor (PEBP1), cell growth suppressors (RASSF5A), cytokine suppressors (SOCS4), transcription factors (SOX17, TCF21) and gastric mucosa protection (TFF1). Interestingly, studies which looked at both gene expression and protein expression, used reverse transcription-polymerase chain reaction (RT-PCR) to amplify up mRNA levels for gene expression analysis. The authors of the review concluded that despite these many genes being identified that there is still a long way to go before any potential clinical use due to the high degree of individual variation between patients and there is still a lack of large scale studies.
Colorectal cancer and B cell lymphomas have previously successfully had a panel of genes identified which can aid in the prognosis of the two diseases (10,11). This has been achieved by tissue analysis with microarray analysis [specifically quantitative nuclease protection assays (qNPA)]. NPA technology analyses mRNA levels without the need for initial extraction followed by RT-PCR, which allows the use of formalin fixed paraffin-embedded (FFPE) tissues. The benefit of this assay over RT-PCR is that it allows the simultaneous processing of large sample numbers with only small amounts of preserved tissue—essential where only biopsies of tumour tissue may be available. It also reduces the amount of pre-processing that would be necessary for RT-PCR analysis.
Using this microarray technique, we studied the expression of 32 candidate genes in historical GC specimens and then looked to verify the expression levels using immunohistochemistry (IHC) analysis. These 32 genes were identified as having potential prognostic interest or value in GC. Our aim was to determine if any yet unidentified genes could be used as an aid in the assessment of prognosis for GC patients. To our knowledge, although previous studies have used RT-PCR to assess gene expression and then followed this with immunohistochemical analysis, this is the first study to utilise microarray technology to look at gene expression in GC and then attempt verification using immunohistochemistry.
Methods
Patients
The University Hospital Coventry and Warwickshire (UHCW) pathology database was searched for all gastric adenocarcinomas diagnosed between April 2005 and September 2006. Demographic data was recorded detailing the patients’ age, sex, tumour stage and survival. As the records were historical there was little data available on smoking status etc. Fifty-seven patients were identified and divided into two groups, those with metastatic disease (n=22), those with no metastatic disease (n=35), who went on to have either curative intent surgery, or who did not have surgery for various reasons, including frailty and or mortality or morbidity. Ethical approval was obtained from the Birmingham Research Ethics Committee, the West Midlands (10/H1210/9).
Gene selection
We selected a panel of 32 genes, some of which have been previously reported as having prognostic value in GC (e.g., E-cadherin), others were selected as they have been shown to have prognostic value in other cancer (BCAS1). Housekeeping genes were incorporated by High Throughput Genomics (HTG) (Inc., Tucson, AZ, USA) genomics (GAPDH, PPIA, RPLPO, ACTB) and were used as a reference set and enabled normalization of data between samples. The full list of genes is shown in Table 1.
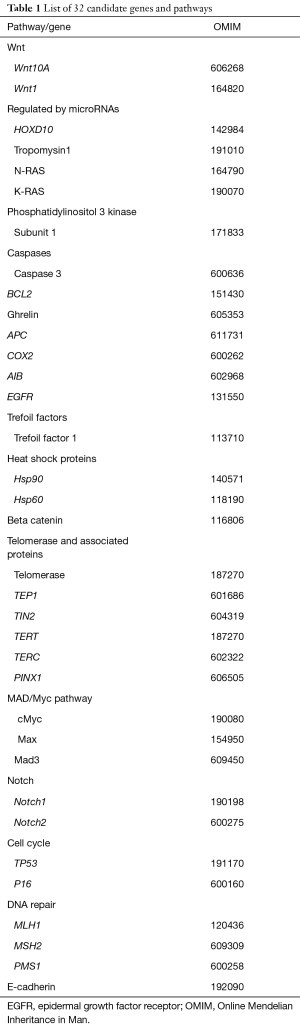
Full table
Preparation of FFPE tissue specimens
Historical tissue specimens were selected and recovered from the tissue archives. The specimens had previously been fixed in FFPE.
Sample preparation for ArrayPlate assay
FFPE sections were prepared and analysed using qNPA ArrayPlates by High Throughput Genomics, Inc. (Tucson, AZ, USA). The FFPE specimens were cut to 5-mm thickness and placed into 75 mL HTG lysis buffer (25 mL/section), vortexed briefly, then heated at 95 °C for 10 min, re-vortexed briefly, and finally frozen at −70 °C until analysis. One cut of 5-mm-thick tissue was used per well on the ArrayPlate, but the HTG lysis buffer containing the multiple sections was used for the three ArrayPlate wells required to measure all the genes in the assay, or for replication in the validation assays. Roberts et al. found in their work that unless the FFPE tissue was thinly cut then poor results were achieved (10).
ArrayPlate assay
To confirm the expression levels of the target genes, qNPA was performed on RNA extracted from FFPE GC sections. The DNA probes for the genes of interest were incubated with the processed tumour samples forming probe-mRNA complexes. Unhybridized probes were then digested by S1 nuclease. Alkaline hydrolysis was then used to destroy the mRNA in the duplexes leaving only intact probes at concentrations reflective of the amounts of original mRNA present. The processed samples were transferred to programmed (linker-modified) ArrayPlates. The ArrayPlates were then exposed to linker oligonucleotides to bind allow specific binding to the probes. The ArrayPlates were then washed with detection oligonucleotides which bound to the linker molecules. The detection oligonucleotides contained horseradish peroxidase, which upon addition of a chemiluminescent peroxidase substrate led to each array element giving off a light signal in proportion to the amount of sample probe bound to the well at that position.
The signals from the ArrayPlates were viewed from the bottom with an OMIX HD imager. The digital images of ArrayPlates were analyzed by ArrayPlate Fit (v.3.31a) software. The resulting data were analyzed by ArrayPlate Crunch software to normalize signals with housekeeping genes and to calculate individual gene expression levels. The ArrayPlate assay has been more extensively described previously by Roberts et al. (10).
Immunohistochemistry
Once gene expression levels were known for the various genes in the GC FFPE specimens, Standard immunohistochemistry techniques were performed on the original FFPE tissue using commercially purchased antibodies against the proteins encoded by the various elected genes. The FFPE specimens were sliced, washed, the primary antibody annealed to them, then rewashed and the secondary antibody applied and rewashed. The stained specimens were then reviewed by two consultant pathologists, with the second blinded, to assess the levels of protein expression for the various identified genes.
Statistical methods
We used a multivariate Cox proportional hazard model. It was fit with stepwise factor selection to determine a set of most relevant factors for the length of survival. Bayesian information criterion (BIC) was used to select the factors ensuring accurate fit while punishing against the inclusion of unnecessary many variables. Apart from the (continuous) gene expression variables, categorical variables for metastatic groups, palliative status, as well as basic patient information (age, sex) was included in the set up.
Two main challenges from the data set had to be taken into account. Firstly, the number of patients is small compared to the number of factors considered. Rather than interpreting significance levels in the traditional univariate sense we understand them as ranks providing a list of most relevant factors. Secondly, gene measurements were taken from different types of samples (only tumour, only biopsy, both). However, there is validity within patients and justifying that both sample types are meaningful. For the analysis, biopsy samples were used whenever available. Analysis was performed with open access software R and packages survival and MASS.
Results
The demographic data of the GC patients are shown in Table 2. The mean age was 72 years [standard deviation (SD): 11.5] and 39 (68%) were male. The mean age of the non-metastatic group was 71 (SD: 13) and the mean age of the metastatic group was 73.5 (SD: 9). The mean survival time of non-metastatic was 23.6 months (SD: 26.4) and the mean survival of metastatic was 7.0 months (SD: 10.0). As expected the mean survival time was highly dependent on metastatic status.
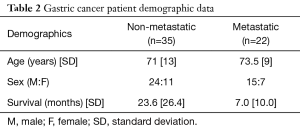
Full table
Gene expression
Stepwise model selection using multivariate Cox PH model including metastatic status, age, sex and 32 gene expression values returned an optimal model which showed that metastatic status, age, sex and five genes appeared to influence the survival in GC. Genes which appears to negatively influence survival (i.e., shorten) include; BCAS1, P53 and HSP90AA1 with relative risks [exp(coef)] of 2.20, 3.73 and 7.53 respectively. Genes which appeared to convey a survival benefit on patients included CASP3 and to a lesser extent, TERT with relative risks of 0.10 and 0.24 respectively. A list of these genes, the Cox PH survival coefficients and p values can be found in Table 3.
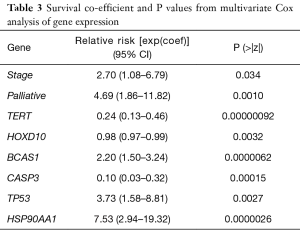
Full table
Immunohistochemical analysis
IHC analysis of the GC tissue specimens was carried out as described above. Genes which were stained for included the discriminating genes P53, HSP90AA1, CASP3 and BCAS1, and the non-discriminating genes NOTCH1, MLH, PSM1 and HOXD10 as controls. Nine non-metastatic patients (i.e., had curative intent surgery) and nine metastatic patients had their tissue samples subjected to IHC assessment as described in the methods. The resulting stained specimens were analysed by two Consultant pathologists (the second blinded) and scored for their expression of levels. The two groups of patients were age and sex matched to each other for analysis.
The CASP3 staining failed and there was insufficient tissue on several samples such that repeat attempts at staining would not have allowed for meaningful comparison. Similarly, two of the patients’ samples were unsuitable for analysis after processing and staining. Which left seven age sex matched patient specimens for comparison (Table 4).
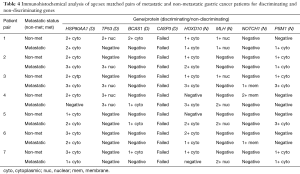
Full table
The results of the IHC staining did not verify the results of the microarray analysis. The expression levels of proteins were either the same in non-metastatic and metastatic specimens or the expression was reduced in the metastatic specimens for genes which carried a negative survival benefit in the microarray analysis. In the case of HSP90AA1 the expression levels were less in the metastatic specimen in three out of seven pairs, the same in one out of seven pairs and more in three out seven pairs. TP53 had lower levels of expression metastatic specimens compared to non-metastatic specimens in one out of seven pairs, higher levels similar levels in two out seven pairs and equal levels of expression in four out of seven pairs. BCAS1 showed similar levels of expression in three out of seven pairs, reduced expression in metastatic specimens in two out of seven pairs and increased expression in metastatic specimens for two out of seven pairs.
The non-discriminating genes, which showed no effect on survival, gave a very similar mixed pattern of similar, reduced and increased expression in metastatic specimens.
The pathologist interpretation of the IHC staining was in absolute or close agreement in 90% of stained samples. The remaining samples where there was a larger discrepancy did not affect the pattern of IHC staining described above.
Discussion
Our study sought to determine whether any yet unidentified genes could be identified to assist with determining prognosis in GC. This expands on previous studies which have identified many genes which appear to affect the survival of GC patients. Statistical analysis of our gene expression data, based on qNPA technology, identified that prognosis was, unsurprisingly, negatively affected by increasing age, male sex and metastatic disease. It also identified five genes (which have statistically significant P values) which affect GC prognosis: three negatively i.e., poorer survival (BCAS1, P53 and HSP90AA1) and two positively i.e., better survival (CASP3 and TERT). Four of these discriminating genes were analysed by IHC techniques in seven non-metastatic patients who had gastric resections with curative intent, and seven age and sex matched metastatic patients. The IHC results showed that the protein expression levels within the tissue samples did not correspond with the mRNA gene expression levels determined in the NPA microassays. This appeared to be the case not just for the discriminating genes that were chosen but also for the non-discriminating genes (NOTCH1, HOXD10, PSM1 and MLH).
Whilst our study appears to have identified several genes which at an mRNA levels may affect the prognosis of GC, the protein expression of these genes appears to be uncoupled from the gene expression. The precise point of the protein expression pathway (i.e., translation, post translational modification) at which this uncoupling process occurs could not be determined from this experiment. The mechanisms behind why this uncoupling has occurred will require further study but they may represent part of the more generalised cell malfunction which occurs with carcinogenesis.
Interestingly this is not the first study to demonstrate that protein expression appears to be disparate from gene expression in cancerous cells. Dickson et al. demonstrated that JAG1 gene and protein expression in breast cancer is associated with poorer prognosis, however, when they carried out IHC studies they determined that there was only a 65% agreement between mRNA and protein levels (12). Stark et al. studied protein and mRNA levels in primary breast cancer and in brain metastases. They demonstrated that whilst BCL-2 mRNA and protein expression levels were in good agreement (and lower in the brain metastases), P53 mRNA levels were significantly lower in the metastases than the primary tumours but the protein levels were only slightly lower in the metastases (not to significant levels). Finally, they found that BAX mRNA and protein levels were completely discordant with the metastases showing lower levels of mRNA expression than the primary tumour, but higher levels of protein expression (13). Sarro et al. studied CD20 levels in chronic lymphocytic leukemia (CLL) and demonstrated that CD20 mRNA levels where normal or near normal compared to healthy controls whilst the CD20 protein levels were reduced by ~60% in CLL cells compared to healthy controls (14). Finally, it has been demonstrated in prostate cancer that mRNA and protein levels of MMP-2, MMP-9 and TIMP-1 show no significant correlation (15).
The previous studies using microarray technology and immunohistochemistry to assess gene expression in colorectal cancer and B cell lymphoma found that protein expression levels did correlate with the microarray findings (11,12). This could suggest that GC has a different cellular behaviour to the other cancers previously studied using this method, or that the genes we selected were not fundamental to the GC pathogenesis process.
Conclusions
Our study utilised microarray technology to try to identify potential gene candidates to aid in determining the prognosis of GC. Biopsy specimens were used in most cases to make a prognostic assignment possible using endoscopic biopsies take at initial cancer diagnosis. We identified five potential genes utilising the microarray technique on FFPE specimens. We then undertook subsequent IHC analysis of the identified genes. This is to our knowledge the first time this has been done on GC specimens. The IHC analysis did not show concordance in the mRNA levels between either the discriminating genes or the control genes we selected. This suggests that there is a disconnection between the gene expression and protein expression of GC cells. Given this finding it is too early to suggest whether these identified genes could have roles as prognostic biomarkers, or as predictors of response to therapy. Further, larger studies, including a verification cohort would be necessary to determine if the mRNA and protein expression findings of our study are a true reflection of the cellular processes which occur during GC carcinogenesis. The quest for a biomarker to aid in the diagnosis and prognosis of GC continues.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the regional West Midlands—Birmingham research ethics committee (10/H1210/9). Written informed consent was waived, as this study used historic histology samples.
References
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol 2006;12:354-62. [Crossref] [PubMed]
- Bilici A. Treatment options in patients with metastatic gastric cancer:current status and future perspectives. World J Gastroenterol 2014;20:3905-15. [Crossref] [PubMed]
- Field K, Michael M, Leong T. Locally advanced and metastatic gastric cancer:current management and new treatment developments. Drugs 2008;68:299-317. [Crossref] [PubMed]
- Wagner AD, Unverzagt S, Grothe W, et al. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev 2010.CD004064. [PubMed]
- Chen Y, Awan N, Haveman JW, et al. Gastric cancer: Australian outcomes of multi-modality treatment with curative intent. ANZ J Surg 2016;86:386-90. [Crossref] [PubMed]
- Wang YX, Shao QS, Yang Q, et al. Clinicopathological characteristics and prognosis of early gastric cancer after gastrectomy. Chin Med J 2012;125:770-4. [PubMed]
- Kanda M, Kodera Y. Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol 2015;21:9838-52. [Crossref] [PubMed]
- Guilford P, Hopkins J, Harraway J, et al. E-cadherin germline mutations in familial gastric cancer. Nature 1998;392:402-5. [Crossref] [PubMed]
- Allgayer H, Babic R, Gruetzner KU, et al. c-erbB-2 is of independent prognostic relevance in gastric cancer and is associated with the expression of tumor-associated protease systems. J Clin Oncol 2000;18:2201-9. [Crossref] [PubMed]
- Roberts RA, Sabalos CM, LeBlanc ML, et al. Quantitative nuclease protection assay in paraffin-embedded tissue replicates prognostic microarray gene expression in diffuse large-B-cell lymphoma. Lab Invest 2007;87:979-97. [Crossref] [PubMed]
- Katkoori VR, Shanmugam C, Jia X, et al. Prognostic significance and gene expression profiles of p53 mutations in microsatellite-stable stage III colorectal adenocarcinomas. PLoS One 2012;7:e30020. [Crossref] [PubMed]
- Dickson BC, Mulligan AM, Zhang H, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol 2007;20:685-93. [Crossref] [PubMed]
- Stark AM, Pfannenschmidt S, Tscheslog H, et al. Reduced mRNA and protein expression of BCL-2 versus decreased mRNA and increased protein expression of BAX in breast cancer brain metastases: a real-time PCR and immunohistochemical evaluation. Neurol Res 2006;28:787-93. [Crossref] [PubMed]
- Sarro SM, Unruh TL, Zuccolo J, et al. Quantification of CD20 mRNA and protein levels in chronic lymphocytic leukemia suggests a post-transcriptional defect. Leuk Res 2010;34:1670-3. [Crossref] [PubMed]
- Lichtinghagen R, Musholt PB, Lein M, et al. Different mRNA and protein expression of matrix metalloproteinases 2 and 9 and tissue inhibitor of metalloproteinases 1 in benign and malignant prostate tissue. Eur Urol 2002;42:398-406. [Crossref] [PubMed]


