
Expression of miR-520c in intestinal type gastric adenocarcinoma
Introduction
Gastric cancer is the most common cancer of digestive tract and the fourth prevalent cancer worldwide (1). As the most lethal global malignant neoplasm after lung cancer (2), gastric adenocarcinoma is gaining more break-out among men especially in developing countries. However, this malignancy has hit second rank among Iranian men (3). Among several histological types of gastric cancer, intestinal-type gastric adenocarcinoma (IGA) is the more prevalent type of gastric malignancies (4). As the preliminary stages of gastric cancer are asymptomatic, early diagnosis in most cases is difficult. Abdominal pain, weight loss and nausea are the usual signs of progressive tumor (5). Currently, several treatment modalities including chemotherapy, radiotherapy and gastrectomy are applied for patients however, the cure and survival of patients are not satisfactory (6).
Increasing evidences represent link between gastric cancer and tens of oncogenes and tumor-suppressors. Although disorders in cellular mechanisms including cell cycle, DNA repair, apoptosis, cell adhesion, differentiation and epigenetic dysregulation have been reported in this multi-gene related malignancy (7) however, exact molecular pathology remains unclear.
Recent studies have reported link between cancer and novel aspect of post-translational gene regulation. MicroRNAs are small single-stranded, non-coding RNAs ranging 18 to 24 nucleotides in length and participate in post-translational gene regulation in eukaryotes via base pairing to their target’s 3’-untranslated region that results in degradation or translational repression of their target mRNAs. More than 2,500 mature miRNAs have been known in human and many are to be validated (available at http://www.mirbase.org/). miRNAs are known to involve in many cellular functions, including proliferation, apoptosis and differentiation (8). miRNAs may play tumor suppressor or oncogenic role in many cancer types. As reported in literatures, many of them are down-regulated or up-regulated in malignancies. Some miRNAs called metastamiRs are known to be involved in metastasis too (9).
miRNAs have been shown to be dysregulated in gastric cancer too. Many miRNAs have been described to be overexpressed or underexpressed in gastric cancer and many others are correlated with gastric cancer prognosis. Some are designated as circulating miRNAs in serum as markers for gastric cancer (10).
miR-520c is a member of miR-515 gene family. miR-520c gene is located on chromosome 19 and encodes an 87-base long precursor miRNA that later would be processed to two mature miRNA called hsa-miR520c-3p and has-miR-520c-5p. hsa-miR-520c-3p that we would call it miR-520c in this article (its previous ID), is a 22-base long miRNA processed from 3’ arm of its precursor, has been characterized as an oncogenic miRNA in several studies.
In this study we analyzed miR-520c probable divergent expression in IGA in 42 pairs of endoscopic samples compared with adjacent non-tumorous tissues from Azeri patients to examine its putative role as oncomiR and its eventual biomarker value in IGA.
Methods
Patients and biopsies
Tumor samples and adjacent non-tumorous tissues were obtained from 48 people referred to Imam-Reza Hospital and Shahid-Madani Hospital in Tabriz, Iran for endoscopy from November 2013 to December 2015. Among them, 42 patients were diagnosed with IGA and selected for study. Patients often were suffering from abdominal pain, weight loss and nausea and had not received any previous medication, chemotherapy or radiotherapy. Endoscopy was performed by specialist physician and the samples were transferred to RNase free micro-tube. The samples were snap frozen in liquid nitrogen and transferred to −80 °C freezer for later RNA extraction. Also, corresponding samples were immediately fixed in 10% formalin for 24 hours and dehydrating, paraffin-embedding, microdissecting into 5 µm tick sections, and staining with H&E were done respectively to samples be diagnosed histologically according to Lauren’s classification and tumor grade (well-differentiated, moderately-differentiated and poorly-differentiated) (11). Since the samples were endoscopy biopsies, the information for tumor size, lymph node involvement, depth of invasion and distant metastasis were not obtained.
Informed written consent was obtained from patients and Medical Ethics Committee of Imam-Reza Hospital approved the study (medical ethics number: tbzmed.irec.1394.517).
Total RNA preparation
Total RNA was isolated from tumor tissues and corresponding adjacent non-tumorous tissues using Trizol reagent (Invitrogen) according to the manufacturer’s instructions with some modification. So that, after adding isopropanol, the solution was incubated for one hour in room temperature instead of 10 min to better precipitate miRNAs. Thirty µL of DEPC-treated water was added to RNA pellet and incubated in water bath at 55 °C for 15 min after pipetting through sampler tip. RNA quality and concentration were assessed using Picodrop Spectrophotometer (Picodrop Ltd.).
Quantification of miR-520c
RNA was first treated with DNase I enzyme to digest any probable DNA remained from RNA extraction procedure (12). The 10 µL treatment reaction contained 2 µg RNA, 1 µL/10× DNase I buffer, 1 µL (1 U) Thermo Scientific DNase I, RNase-free enzyme, 0.5 µL RiboLock RNase Inhibitor enzyme and DEPC-treated water. The mixture was incubated at 37 °C for 30 min in 96-well Biometra thermocycler. To inactivate DNase I enzyme, we added 1 µL of 50 mM EDTA and incubated the mixture at 65 °C for 10 min (13).
Prior to miRNA quantification, the DNase I treated RNA was 3'-extended with poly-A tail using poly-A polymerase enzyme. The 20 µL reaction mixture contained 2 µg RNA (10 µL total volume from previous step), 2 µL/10× reaction buffer, 2 µL/10 mM ATP and 0.5 µL poly-A polymerase enzyme. The reaction was incubated for 10 min at 37 °C.
Real-time revers transcription polymerase chain reaction (qRT-PCR) was performed using PARSGENOME MiR-Amp kit. Briefly, the miR-520c and U6 (as internal control) was reverse-transcribed from total RNA using specific primers. The 10 µL RT reaction mixture contained 2 µL/5× RT reaction buffer, 1 µL/10 mM dNTP mix, 0.5 µL RT enzyme, 0.5 µL each specific primers for miR-520c and U6 as internal control and 5.5 µL poly-A extended RNA. The reaction condition was as follows: incubation for 60 min at 43 °C and incubation for 1 min at 85 °C to inactivate the RT enzyme. The synthesized cDNA was diluted up to 2 times and stored at −80 °C for subsequent real-time PCR.
The relative real-time PCR quantification was performed on QIAGEN Rotor-Gene Q system using SYBER Green Real-Time PCR Master Mix. The 20 µL reaction mixture includes 10 µL SYBER Green Real-Time PCR Master Mix, 1 µL mix of specific forward primer and universal reverse primer, 8 µL ddH2O and 1 µL RT product of total RNA. The reaction conditions were as follows: Initial denaturation at 95 °C for 1 min followed by 50 cycles of denaturation at 95 °C for 5 s, annealing at 61 °C for 15 s and extension at 72 °C for 30 s followed by final extension for 2 min at 72 °C. All samples were processed in duplicate. Duplicate samples differing on Ct value more than 0.4 were repeated. Ct values were detected by thermocycler as the cycle at which the fluorescence absorbed by system passed the threshold. Standard curve was plotted for both miR-520c and U6 using serial dilutions of cDNA as 1, 0.2, 0.04, 0.008 and 0.0016. For each real-time PCR, melting curve was generated to monitor reaction specificity.
Data analysis
Relative expression ratio between tumor samples and adjacent non-tumorous tissues was analyzed using both Pfaffl method and delta-delta Ct method (ΔΔCt).
For Pfaffl method we applied REST 2009 Software Tool V2.0.13 (QIAGEN Gmbh). This software utilizes Pfaffl equation (14) as follows:
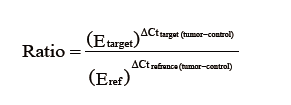
For ΔΔCt method (15), Paired-samples t-test was employed to compare the difference between mean expression of miR-520c in tumor and non-tumorous tissues. ΔCttarget was calculated as Ctmir – CtU6 for tumor samples. ΔCtnormal was also calculated for non-tumorous tissues as Ctmir – CtU6. Then the normalized miR-520c expression was calculated separately for tumor tissue and adjacent normal samples using equation:
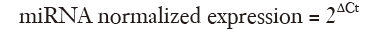
The resulting values were the input data for SPSS software.
Moreover, statistical analysis between groups were analyzed using SPSS® 16.0 (SPSS Inc.) for Windows®. To analyze correlation between miR-520c expression and clinicopathological features, Spearman correlation coefficient was applied. Kolmogorov-Simonov test was done to examine whether the expression of miR-520c is normal among samples. To analyze the homogeneity of variances Leven test was performed.
Results
We evaluated the expression levels of miR-520c in 42 pairs of IGA tissues and adjacent non-tumorous tissues by real-time revers transcription PCR. The sharp melting-curves of miR-520c and housekeeping gene U6 and a single band on agarose gel electrophoresis of the qRT-PCR products confirmed the reaction specificity for miR-520c and U6. The analysis results by both REST and SPSS software showed that there is no significant expression change of miR-520c in tumor tissues in comparison to that in the adjacent normal tissues (Figure 1).
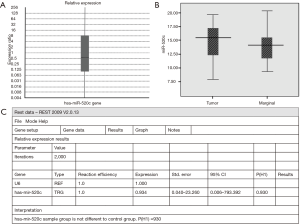
We further determined correlation between miR-520c expression and clinicopathological features of patients in IGA. As shown in Table 1 any significant correlation was found between miR-520c expression and clinicopathological features of patients including gender, tissue grade and age.

Full table
Discussion
Recently, microRNAs have been found to involve in many cellular mechanisms. Their important roles in cell functions indicate their major involvement in pathological conditions. Since discovery of first miRNAs, ambitious researchers have endeavored to uncover this novel aspect of biology.
Along with other pathological conditions, cancer studies have benefited from miRNA discovery. So far, thousands of miRNAs have been detected to involve in cancer onset, development, metastasis and cancer relapse. These miRNAs misexpression may be as a result of their genes failure, like aberrant methylation of their promoter (16) and mutation in their genes (17), or misexpression may be as a consequence of other oncogenes or tumor-suppressors (18). Many miRNAs have been shown to down-regulate or up-regulate in certain types of cancers (19,20). This fact has opened another field in cancer-therapy studies (21) and elevated hopes to find more useful biomarkers for cancer prognosis and diagnosis (22).
Gastric cancer, as the second lethal cancer worldwide, is receiving more attention in miRNA related studies. Aberrant expression of many miRNAs has been detected in gastric cancer (23,24) and several miRNAs have been identified as biomarkers for this malignancy (25,26). More importantly, recent studies have identified miRNAs as non-invasive biomarker candidates in gastric cancer. Cai et al. identified that plasma miRNAs of miR-106b, miR-20a, and miR-221, may act as potential non-invasive biomarkers for early detection of gastric cancer (27). Also, Tsujiura et al. showed that plasma concentration of miR-17-5p, miR-21, miR-106a, miR-106b were significantly higher in gastric cancer patients than controls (28).
Several studies have proposed miR-520c as potential oncogene and metastasis driven microRNA that exerts its functions by targeting tumor suppressors. Huang et al. in a study applied trans-well migration assay to find out miRNAs with metastasis promoting potency using non-metastatic MCF-7 cell line that was transduced with a miRNA-expression library (29). They found that human miR-373 and miR-520c can stimulate cancer cell migration and invasion in vitro and in vivo by targeting 3'UTR region of CD44 gene. Other study by Yang et al. showed that miR-520c and miR-373 exert pro-invasive effects in PC-3M cells by suppressing CD44 in protein level by binding its 3'UTR region (30). Liu and Wilson in another study explored miR-520c and miR-373 effects on MMP9 mRNA in human fibrosarcoma HT1080 cells. They described that these miRNAs upregulate MMP9 expression, translation and activity by directly targeting the 3'UTR of mRNAs of mTOR and SIRT1, negative regulators of expression of MMP9 via inactivating the Ras/Raf/MEK/Erk signaling pathway and transcription factor NF-κB activity and, thus suppressing translation levels of SIRT1 and mTOR that ends in enhancing cell migration and cell growth in 3D type I collagen gels (31).
In this study, we explored expression of miR-520c in IGA, the major malignancy of stomach, to find out whether this miRNA plays mentioned oncogenic role and also to evaluate its value as biomarker for IGA detection. Our study on 42 pairs tissue samples of IGA and adjacent normal tissues showed any significant correlation between IGA and miRNA-520c, a finding that opposes previous findings of this miRNA. This finding suggests conflicting roles for miRNAs and different malignancy mechanisms in cancer. Since different molecular aberrations may result in gastric cancer, it is probable that miRNA-520c drives its oncogenic character via distinct procedure or in a specific stage of tumor formation and invasion as the above-mentioned studies concentrated on miR-520c as MetastamiR and invasion promoting microRNA.
Conclusions
All in all, our study resulted in conflicted role for miR-520c as an oncogene and denies oncogenic role for miR-520c in IGA and its capability as biomarker in gastric cancer.
Acknowledgements
We thank the personnel of Endoscopy Department of Shahid-Madani Hospital and Imam-Reza Hospital specially, Mrs. Shahin Fotovat and Mrs. Fariba Farhangmehr for their help to gather samples and Mr. Farhad Nazari for his efforts in preparing samples for histological analysis.
Funding: This work was supported by Liver and Gastrointestinal Disease Research Center of Tabriz University of Medical Sciences (grant number 137/219).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Medical Ethics Committee of Imam-Reza Hospital approved the study (medical ethics number: tbzmed.irec.1394.517) and informed written consent was obtained from patients.
References
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Parkin DM. International variation. Oncogene 2004;23:6329-40. [Crossref] [PubMed]
- Aghjani, H, Etemad K, Gouya M, et al. Iranian Annual of National Cancer Registration Report 2008-2009 Ministry of Health & Medical Education. Center for Disease Control & Prevention Noncommunicable Diseases Unit Cancer Office, Technical Report 2011.
- Polkowski W, van Sandick JW, Offerhaus GJ, et al. Prognostic value of Laurén classification and c-erbB-2 oncogene overexpression in adenocarcinoma of the esophagus and gastroesophageal junction. Ann Surg Oncol 1999;6:290-7. [Crossref] [PubMed]
- Layke JC, Lopez PP. Gastric cancer: diagnosis and treatment options. Am Fam Physician 2004;69:1133-40. [PubMed]
- Takahashi T, Saikawa Y, Kitagawa Y. Gastric Cancer: Current Status of Diagnosis and Treatment. Cancers 2013;5:48-63. [Crossref] [PubMed]
- Nobili S, Bruno L, Landini I, et al. Genomic and genetic alterations influence the progression of gastric cancer. World J Gastroenterol 2011;17:290-9. [Crossref] [PubMed]
- He L, Gregory J. Hannon. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Hurst DR, Edmonds MD, Welch DR. Metastamir: The Field of Metastasis Regulatory microRNA is Spreading. Cancer Res 2009;69:7495. [Crossref] [PubMed]
- Yin Y, Li J, Chen S, et al. MicroRNAs as Diagnostic Biomarkers in Gastric Cancer. Int J Mol Sci 2012;13:12544-55. [Crossref] [PubMed]
- Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand 1965;64:31-9. [Crossref] [PubMed]
- Kienzle N, Young D, Zehntner S, et al. DNase I treatment is a prerequisite for the amplification of cDNA from episomal-based genes. BioTechniques 1996;20:612-6. [PubMed]
- Wiame I, Remy S, Swennen R, et al. Irreversible heat inactivation of DNase I without RNA degradation. BioTechniques 2000;29:252-4. [Crossref] [PubMed]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [Crossref] [PubMed]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 2008;3:1101-8. [Crossref] [PubMed]
- Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv Clin Chem 2010;52:145-67. [Crossref] [PubMed]
- Kotani A, Ha D, Schotte D, et al. A novel mutation in miR-128b gene reduces miRNA processing and leads to Glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell cycle 2010;9:1037-42. [Crossref] [PubMed]
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 2005;353:1768-71. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Ahmad J, Hasnain SE, Siddiqui MA, et al. MicroRNA in carcinogenesis & cancer diagnostics: A new paradigm. Indian J Med Res 2013;137:680-94. [PubMed]
- Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res 2011;157:216-25. [Crossref] [PubMed]
- Heneghan HM, Miller N, Kerin MJ. MiRNAs as biomarkers and therapeutic targets in cancer. Curr Opin Pharmacol 2010;10:543-50. [Crossref] [PubMed]
- Tang GH, Tang M, Xie YJ. The Role of miRNAs in Gastric Cancer. J Gastroint Dig Syst 2013;3:129. [Crossref]
- Pan HW, Li SC, Tsai KW. MicroRNA dysregulation in gastric cancer. Curr Pharm Des 2013;19:1273-84. [PubMed]
- Wang F, Sun GP, Zou YF, et al. MicroRNAs as promising biomarkers for gastric cancer. Cancer Biomark 2012;11:259-67. [Crossref] [PubMed]
- Wang JL, Hu Y, Kong X, et al. Candidate microRNA biomarkers in human gastric cancer: a systematic review and validation study. PLoS One 2013;8:e73683. [Crossref] [PubMed]
- Cai H, Yuan Y, Hao Y, et al. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol 2013;30:452. [Crossref] [PubMed]
- Tsujiura M, Ichikawa D, Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 2010;102:1174-9. [Crossref] [PubMed]
- Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 2008;10:202-10. [Crossref] [PubMed]
- Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol 2009;2:361-9. [PubMed]
- Liu P, Wilson MJ. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-κB factor in human fibrosarcoma cells. J Cell Physiol 2012;227:867-76. [Crossref] [PubMed]


