Investigation of the changes in the expression levels of MOZ gene in colorectal cancer tissues
Introduction
Colorectal cancer (CRC) is one of the most important causes of mortality in the world and includes 9% of total occurred cancers. That is third most common cancer and forth cause of death in the world (1). Different studies revealed that this cancer began to expansion in Iranian population recently. Cause of its expansion may be related to alteration in lifestyle, smoking, reducing physical activity, and malnutrition. About 35% of the risk for cancer relates to genes (2). Other than the genetical cause, a lot of evidences have showed that inappropriate epigenetic changes have an important role in human cancers. Many DNA methylation and histone modifications have been used as a cancer biomarker. Because of reversible state of the epigenetic modification, much therapeutics have been adopted on this basis (3). HATs and histone deacetylases (HDAC) are responsible for adding and separating acetyl group for lysine residue of histones, respectively. Losing balance of HATs and HDACs in tumorous cells causes inactivating of tumor suppressor genes transcription (4). HATs are various groups of enzymes that based on their catalytic domain are divided into different groups (5). MOZ (KAT6A) is one of the most important HAT enzymes that belongs to MYST family (6). This gene has been located on chromosome 8 breakpoint that encodes protein containing 2004 amino-acids, and characterizing by two C4HC3 zinc fingers and a single C2HC zinc finger domain which they have a role in acetylation. The t(8;16) (p11;p13) translocation, which associates with acute myeloid leukemia (AML), fuses the MOZ gene on 8p11 with CBP gene on 16p13 (7), thereby causes enhanced cell proliferation. It has an important role in acetylation of lysine 9 residue in histone 3. MOZ by acetylating p53, performs its role in p53 signaling pathway (8). These studies have showed that MOZ has dual property in hematological malignancies. Therefore, contribution of MOZ in human leukemia was studied, extensively (9-11). The role of MOZ expression levels in solid tumors is less revealed. On the other hand, it is necessary to evaluate its role in solid tumors that are prevalent. Extensive researches on molecular biology have used qRT-PCR as an important technique to analysis mRNA expression pattern and compare the relative levels of mRNA between normal and tumorous tissues (12). On this basis, we decided to analysis MOZ gene expression using qRT-PCR technique in CRC that is widespread in the northwest of Iran and compare with other similar studies.
Methods
Tissue collection
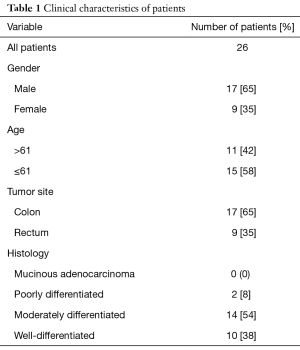
In the present cohort study, tumor tissues and tumor adjacent normal tissues were collected from 26 patients (17 men and 9 women) that referred to Imam Reza and Shahid Madani hospitals of Tabriz, Iran, between September 2016 to February 2017. Paired adjacent normal appearing tissues were taken at distance ~10 cm from the tumor position. The biopsies were snap-frozen in liquid nitrogen and stored in −80 °C freezer until RNA extraction. In pathology laboratory, pathological analysis was performed on tumor samples and surgical records were reviewed for the position of the tumors. None of the patient’s tissues had been received any chemotherapy or other kinds of remedies. Differentiation grade of tumor tissues was recognized by pathology specialist. After finding confidence from the tumoral feature of tissues, subsequent steps were continued. In order to additional examinations, a number of demographic data (age and sexuality) with regarding ethical consideration was collected. Written informed consents were obtained from all patients. The ethics committee of Tabriz University of Medical Sciences permitted the investigation with an institutional protocol. The work was undertaken and that it conforms to the provisions of in accordance with the Helsinki Declaration. Table 1 shows clinicopathological characteristics of all patients.
Full table
Total RNA extraction and reverse transcription of mRNA
Total RNA extraction from tissues was done using RNX-Plus (CinnaGen), exactly according to the protocol which was recommended by the manufacturer. In order to inactivate RNase enzyme, all of the equipment were treated with 1% DEPC water for 24 hours and then autoclaved (Thermolyne). RNAs were stored at −80 °C freezer until cDNA synthesis. Quality and quantity of extracted RNA were determined using 1% agarose gel electrophoresis and UV spectrophotometer (Picodrop, UK) at the absorbance 260/280. For verification accuracy of RNA extraction, preservation of two bands (18S and 28S rRNA) on agarose gel and absorbance ratios between 1.8 and 2 on spectrophotometer were considered. Before cDNA synthesis, in order to removing DNA contamination probability, 2 µg of extracted total RNA was treated by DNaseI (Fermentase Co. Lithuania). Then cDNA was synthesized using 2 µg of RNA as synthesis template, random hexamer primers and reverse transcriptase (Fermentase Co. Lithuania) on thermal cycler (PeQlab, Germany).
qRT-PCR and statistical analysis
In this study housekeeping gene, GAPDH was used as an endogenous control. Primers of MOZ designed by OLIGO v.7 software and specificity of them was evaluated by NCBI Blast (Table 2). At the next step, a qRT-PCR was used for determination of MOZ gene expression in the specimens on Eco™ Real-Time PCR system. The component of the reaction mixture contains 5 µL SYBR® Green Real-Time PCR Master Mixes (Amplicon), 0.2 µL forward and reverse MOZ/GAPDH gene primers, 3.6 µL H2O and 1 µL target cDNA. The condition for PCR is illustrated in Table 3. For endorsement of PCR results, products of PCR were run on agarose gel. All of the samples were amplified simultaneously and the quantitative data were calculated. Collected data were statistically analyzed (ΔCT and 2−ΔΔCT formulas) by IBM SPSS Statistics 24.0 software and independent t-test. Statistical significance was defined as P≤0.05.

Full table
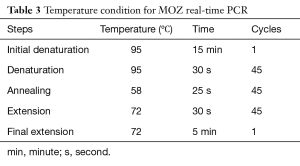
Full table
Results
Clinical characteristics of patients
In this study, 26 patients with CRC include 17 men (65%) and 9 women (35%) with average age 62.35 (range between 38–83) were evaluated. Seventeen had colon cancer and 9 had rectal carcinoma. None of the primary carcinomas were mucinous adenocarcinoma, 2 were poorly, 14 were moderately and 10 were well differentiated (Table 1).
Expression levels of MOZ in colorectal tumor and marginal normal tissues
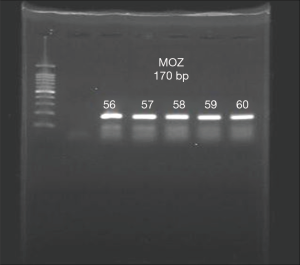
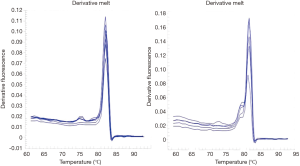
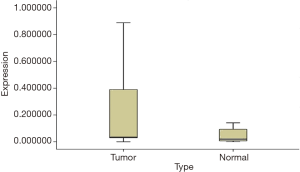
The existence of a band with the desired size (170 bp) and the absence of nonspecific bands to ensure proper synthesis indicated the success of the cDNA synthesis (Figure 1). The annealing temperature for MOZ gene was determined 58 °C (Figure 2). Melt curve of MOZ and GAPDH had one pick that shows one PCR product (Figure 3). In addition, findings of agarose gel endorsed PCR results. The differences in MOZ mRNA expression between CRC and normal tissues were significant. The mean inverse expression ratio of MOZ and GAPDH showed high MOZ levels in CRC compared to normal colorectal tissues (mean expression ratio, 0.180 versus 0.047, P=0.048, CI =95%) (Figure 4).
Correlation between MOZ expression levels and clinicopathological features in patients with CRC
Gene expression levels in different differentiation grades of CRC patients
Analysis showed that there was no significant difference in different differentiation grades (P=0.52).
Gene expression levels in different positions of colorectal tumors
There was no significant difference in different tumor positions (colon and rectum) (P=0.31).
Gene expression levels in two age groups of CRC patients
There was no significant difference in two age groups (≤61 vs. >61) (P=0.69).
Gene expression levels in two sexual groups of CRC patients
There was no significant difference in two sexual groups (male and female) (P=0.37).
Discussion
Multiple histone modifiers and HATs that are misregulated in CRC have been shown by expression evaluation in CRC tissues in comparison with non-cancerous tissues and have been revealed that these groups of epigenetic modifiers are closely associated with CRC (13,14). However, there has been no expression analysis describing the role of MOZ as a HAT gene in CRC. The present study is the first expression evaluation of this HAT on CRC. The present investigation has provided evidence that confirms the contribution of MOZ in malignancy progression based on previous studies. Our research has showed that the level of MOZ mRNA expression between tumor and normal tissues is under P value threshold (0.05), considering as a significant overexpression. Multiple studies have showed that misregulation of MOZ has a key role in tumorigenesis. Besides hematologic malignancies, especially leukemia, recurrent amplification of MOZ has been reported in various solid tumors, including breast cancer, ovarian cancer, uterine cervix cancer, lung adenocarcinoma, colon and rectal cancer (15). Comparative genomic hybridization (CGH) on cDNA microarrays showed hundreds of genes whose overexpression of them is attributable to gene amplification, including MOZ (16). MOZ creates translocation with different genes that whole of them have HAT activity. This gene also has interaction with multiple transcription factors such as transcription activators. Most important translocations of MOZ with transcription factors that participate in AML are MOZ-TIF2, MOZ-CBP (6). In the presence of MOZ, the transcription level of p16 is decreased and consequently allows fast progressing of the cell cycle, but MOZ fusion proteins that create leukemia, such as MOZ-TIF2 suppress the expression of p16, leads to enhanced proliferation and development of leukemia (17). With the other mechanism, MOZ is critical for the expression of multiple repressors of the INK4A/ARF locus by stabilizing the H3K9ac levels at those genes (18). MOZ also is necessary for correct expression of HOX genes (19). In CRC and hepatocellular carcinoma, HOX genes have shown different patterns of expression in comparison with normal tissues (20). Overexpressed MOZ or its fusion states can lead to upregulation of HOX genes and consequently lead to immortalization of progenitor cells in bone marrow. Although, mechanisms that overexpression of MOZ contributes to solid tumorigenesis are not completely clear yet, one possible mechanism is recruitment of TRIM24 to activate PI3KA transcription by MOZ, thereby leads to upregulation of PI3K/ACT signaling and tumor development that has been shown in glioblastoma (GBM) (21). On the bases of these findings, there is a hypothesis that the contribution of MOZ in CRC might be occurred through similar mechanisms. Some researchers have endorsed overexpression of MOZ in some solid tumors. For example, in estrogen receptor+ (ER+) breast cancers (22) as well as luminal breast cancer that show overexpression of MOZ, knock outing this gene induces reducing the proliferation and other symptoms of malignancy. These studies have introduced MOZ as one potential oncogene (23). According to different researches that have reported overexpression of MOZ in tumors and its significant role in cancer, we predict that increasing number of patients and evaluating its expression in massive statistical society in different cancers and especially its expression at protein level will affect research result. Also, the possible contribution of higher expression level of MOZ in tumorigenesis, making it a candidate as a therapeutic target for cancer treatment.
Conclusions
In CRC, we noted significant overexpression at mRNA level of MOZ (P=0.048). These differences in MOZ gene expression in CRC and normal tissues suggested MOZ expression could be a biomarker for screening and diagnosis of CRC in the northwest of Iran.
Acknowledgements
The authors thank the Department of Animal Biology, Faculty of Natural Sciences, University of Tabriz, for providing the research center and laboratory equipment.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The ethics committee of Tabriz University of Medical Sciences permitted the investigation with an institutional protocol. Written informed consents were obtained from all patients.
References
- Haggar FA, Boushey RP. Colorectal Cancer Epidemiology: Incidence, Mortality, Survival, and Risk Factors. Clin Colon Rectal Surg 2009;22:191-7. [Crossref] [PubMed]
- Colorectal Cancer: The Diagnosis and Management of Colorectal Cancer. National Collaborating Centre for Cancer (UK), 2011. (NICE Clinical Guidelines, No. 131.).
- Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457-63. [Crossref] [PubMed]
- Richon VM, Sandhoff TW, Rifkind RA, et al. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A 2000;97:10014-9. [Crossref] [PubMed]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol 2007;8:284-95. [Crossref] [PubMed]
- Katsumoto T, Yoshida N, Kitabayashi I. Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci 2008;99:1523-7. [Crossref] [PubMed]
- Borrow J, Stanton VP Jr, Andresen JM, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 1996;14:33-41. [Crossref] [PubMed]
- Arboleda VA, Lee H, Dorrani N, et al. De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am J Hum Genet 2015;96:498-506. [Crossref] [PubMed]
- Rokudai S, Aikawa Y, Tagata Y, et al. Monocytic leukemia zinc finger (MOZ) interacts with p53 to induce p21 expression and cell-cycle arrest. J Biol Chem 2009;284:237-44. [Crossref] [PubMed]
- Rokudai S, Laptenko O, Arnal SM, et al. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc Natl Acad Sci U S A 2013;110:3895-900. [Crossref] [PubMed]
- Zhu J, Sammons MA, Donahue G, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 2015;525:206-11. [Crossref] [PubMed]
- Jozefczuk J, Adjaye J. Quantitative real-time PCR-based analysis of gene expression. Methods Enzymol 2011;500:99-109. [Crossref] [PubMed]
- Ishihama K, Yamakawa M, Semba S, et al. Expression of HDAC1 and CBP/p300 in human colorectal carcinomas. J Clin Pathol 2007;60:1205-10. [Crossref] [PubMed]
- Huang Fu, Susan M. Abmayr, Jerry L. Workman. Regulation of KAT6 Acetyltransferases and Their Roles in Cell Cycle Progression, Stem Cell Maintenance, and Human Disease. Mol Cell Biol 2016;36:1900-7. [Crossref] [PubMed]
- Kang KA, Piao MJ, Ryu YS, et al. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget 2016;7:40594-620. [Crossref] [PubMed]
- Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 2002;62:6240-5. [PubMed]
- Largeot A, Perez-Campo FM, Marinopoulou E, et al. Expression of the MOZ-TIF2 oncoprotein in mice represses senescence. Exp Hematol 2016;44:231-7.e4. [Crossref] [PubMed]
- Sheikh BN, Phipson B, El-Saafin F, et al. MOZ (MYST3, KAT6A) inhibits senescence via the INK4A-ARF pathway. Oncogene 2015;34:5807-20. [Crossref] [PubMed]
- Voss AK, Collin C, Dixon MP, et al. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell 2009;17:674-86. [Crossref] [PubMed]
- Kanai M, Hamada J, Takada M, et al. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep 2010;23:843-51. [PubMed]
- Lv D, Jia F, Hou Y, et al. Histone Acetyltransferase KAT6A Upregulates PI3K/AKT Signaling through TRIM24 Binding. Cancer Res 2017;77:6190-201. [Crossref] [PubMed]
- Yu L, Liang Y, Cao X, et al. Identification of MYST3 as a novel epigenetic activator of ERα frequently amplified in breast cancer. Oncogene 2017;36:2910-8. [Crossref] [PubMed]
- Turner-Ivey B, Guest ST, Irish JC, et al. KAT6A, a chromatin modifier from the 8p11-p12 amplicon is a candidate oncogene in luminal breast cancer. Neoplasia 2014;16:644-55. [Crossref] [PubMed]