CD133-positive tumor cell content is a predictor of early recurrence in colorectal cancer
Introduction
Although overall survival (OS) has improved substantially in patients with colorectal cancer, the median survival time of patients with metastatic colorectal cancer still does not exceed 2 years. Approximately one-third of patients with stage III colorectal cancer experience tumor recurrence despite aggressive surgery and postoperative chemotherapy. One potential mechanism conferring resistance to chemotherapy is the presence of tumor initiating cells, defined as cells capable of proliferation, self-renewal, and differentiation into various cancer cell types (1). Although the clinical implications of tumor initiating cells have not yet been determined, these cells may be responsible for primary resistance and/or refractoriness to chemotherapy (2).
CD133, a pentaspan transmembrane protein, is a stem cell marker used to identify tumor initiating cells (3), especially in colorectal cancer. CD133 positivity is a marker for putative cancer stem cell populations in the brain (4-6), liver (7), pancreas (8), and colon (9,10). Although CD133+ cells have been shown to be pluripotent, with as few as 1,000 cells capable of tumor initiation (9,10), both CD133-positive and CD133-negative cells have been reported to retain the ability to mediate tumor initiation (11). In vitro cultivation of tumor initiating cells derived from surgical specimens are crucial for the development and screening of novel agents directed against stem cell pathways (2).
We have therefore (I) tested the tumorigenicity of CD133+ cells using LoVo cell line in vitro and in vivo; (II) investigated the proportion of CD133-positive cells isolated from surgical specimens; (III) evaluated the prognostic impact of the percentage of CD133-positive tumor cells on treatment outcome in patients with colorectal cancer; and (IV) established a living tumor tissue bank to screen for various stem cell inhibitors in colorectal cancer.
Materials and methods
Cell culture and reagents
Human colorectal cancer cell line, LoVo, was purchased from Korea Cell Line Bank (KCLB, Seoul, Korea). LoVo cells were maintained in RPMI-1640 medium (PAA Laboratories GmbH, Austria) supplemented with 10% heat-inactivated fetal bovine serum, antibiotic/antimycotic. Cells were incubated at 37 °C in 5% CO2 and the medium changed twice a week. After confluence, cells were subdivided into new flasks until the end of the experiment.
Colony formation assay
About 2×102 cells per well were added into 6-well culture plate with three wells each group. After incubation at 37 °C for 14 days, the cells were washed twice with PBS and stained with 0.5% crystal violet in 20% methanol. The number of colonies was counted under microscope.
Tumor specimens and primary tumor cultures
Human colon cancer tissue specimens were obtained from patients undergoing surgical resection for curative or palliative purposes and processed within 30 minutes of surgical extirpation. The fresh tissue specimens were extensively washed with serum-free RPMI1640, minced, and enzymatically dissociated in serum-free RPMI1640 containing 0.4 mg/mL collagenase (Gibco, Carlsbad, CA, USA), 0.5 mg/mL dispase (Gibco) and 0.2 mg/mL DNase I (Roche, Mannheim, Germany) for 2 h at 37 °C with agitation. Red blood cells were removed by differential centrifugation. A fraction of primary cells from each specimen was cultured in ultra low attachment plates (Corning Inc., Corning, NY, USA) at 37 °C in serum-free CSC medium, consisting of DMEM/F12 medium (Gibco) supplemented with human recombinant bFGF and EGF (20 ng/mL each, PeproTech Inc., Rocky Hill, NJ, USA), 4 μg/mL heparin (Sigma-Aldrich, St Louis, Mo, USA), and B27 (Invitrogen). Another fraction of each cell specimen was cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum.
The study protocol was approved by the Institutional Review Board (IRB) of Samsung Medical Center prior to the collection of tissue specimens, and all patients provided written informed consent.
Fluorescence-activated cell sorting
LoVo cells were detached from the plates with trypsin-EDTA, counted, and washed in PBS containing 0.1% BSA. At least 5×106 cells in 100 μL PBS containing 0.5% BSA were incubated with APC-conjugated fluorescence-labeled mouse anti-human CD133/2 (Miltenyi Biotec, Germany) or APC-conjugated isotype control mouse IgG2b (diluted 1/10) at 4 °C for 30 min in the dark. After washing, the labeled cells were sorted by flow cytometry into CD133+ and CD133– tumor cells using a FACS Aria cell sorter (Becton & Dickinson, Mountain View, CA, USA). The purity of each sorted population was at least 90%.
Flow cytometry assay for CD133 and EpCAM expression on primary tumor cells
Dissociated colorectal cancer cells were washed 3 times with PBS, resuspended in PBS, and incubated with APC-conjugated mouse anti-human CD133/2 (Miltenyi Biotec, Germany) and PE-conjugated mouse anti-human EpCAM (CD326, epithelial cell surface antigen, BD Biosciences, USA). The fluorescence of CD133 and EpCAM was measured and analyzed using a FACS Calibur flow cytometer (BD Biosciences).
Sphere assays
Cells plated at a density of 200 cells/well in 96-well ultra low attachment plates (Corning) in CSC medium were incubated for 14 days. To each well was added 20 μL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenylterazolium bromide (MTT; 5 mg/mL; Promega), and the cells were incubated for 4 h. The spheres were collected by gentle centrifugation at 1,500 rpm, the supernatants were removed, and 150 μL dimethyl sulfoxide (DMSO, Sigma) were added to each well. Fifteen minutes later, the absorbance of each well at 570 nm was measured using a microplate reader. All samples were analyzed in triplicate.
In vivo tumorigenic assay
The animal study protocol was reviewed and approved by the Institutional Animal Care and Use committee (IACUC) of Samsung Biomedical Research Institute. Six-week-old male immune deficient mice, obtained from Orient Laboratory, Korea, were each subcutaneously injected with 2.5×106 CD133+ or CD133– cells, isolated by FACS sorting, in 0.2 mL PBS. The mice were monitored daily, with tumor volume calculated using the formula (a×b2)×0.5, in which “a” is the long axis and “b” is the short axis of the tumor in millimeters. Twenty-four days after injection, all the mice were euthanized, and their tumors were excised aseptically.
Immunohistochemistry
Formalin-fixed paraffin-embedded tissue sections, 3-μm thick, were mounted onto positively charged glass slides. The tissue sections were deparaffinized in xylene, hydrated in descending concentrations of alcohol, and washed in distilled water. The slides were immersed in DAKO High Buffer (pH 8.0) in a 97 °C water bath (PT Link, DAKO) and microwaved twice for 10 min each for antigen retrieval. Following three washes in PBS, the slides were incubated with 3% hydrogen peroxide in distilled water for 5 minutes to block endogenous peroxidases. The slides were incubated with primary antibody to CD133 (clone AC133; diluted 1:50; Miltenyi Biotec, Auburn, CA, Cat No. # 130-090-422) for 60 minutes, washed, incubated with secondary antibody for 25 minutes, washed, and incubated with an DAKO EnvisionTM Detection Kit, Peroxidase/DAB K5007 (DAKO, Glostrup, Denmark) for 50 minutes. After further washing, the slides were incubated in 3% diaminobenzidine for 20 minutes and counterstained with hematoxylin. Dark brown staining of more than 15% of tumor cells was scored as positive.
Statistical analysis
The mean tumor volume in each mouse for each cell line was computed for growth curves (the mean tumor volume in each group = total volume from all mice per group divided by number of mice). Statistical significance of the differences between groups for cell colony formation, sphere formation, and tumor volume was calculated using the Student t-test. Disease-free survival was defined as the time from surgery to documented recurrence or death. The log-rank test was used for univariate analysis and Cox’s regression was used for multivariate analysis. All P values less than 0.05 were considered to be statistically significant. All statistical tests were two-sided.
Results
CD133+ cells as tumorigenic cells in vitro and in vivo
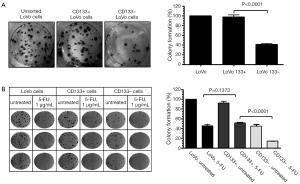
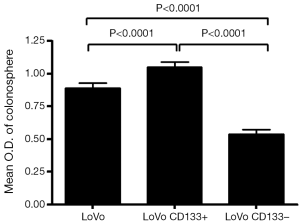
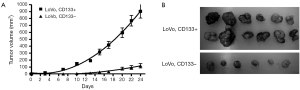
We first tested the tumorigenic potential of CD133+ cells in vitro using clonogenic assay. LoVo cells were sorted into CD133+ and CD133– cells using flow cytometry and were assayed for tumorigenicity in vitro. The CD133+ LoVo cells ranged from 15-33% of the unsorted LoVo cells. As demonstrated in Figure 1A, CD133+ LoVo cells were significantly more tumorigenic than CD133– cells when assessed with clonogenic assay after isolation of CD133+ cells (CD133+ vs. CD133– LoVo cells: mean growth, 93.63% vs. 41.05%; difference, 56.58%; 95% CI, 45.92% to 67.23%; P<0.0001). In addition, CD133+ cells were resistant to 5-fluorouracil by clonogenic assay after 5-FU treatment at 1 µg/mL for 14 days in vitro (Figure 1B). In contrast, unsorted total LoVo cells and CD133– LoVo cells were sensitive to 5-FU (Figure 1B). To further validate tumorigenicity of CD133+ LoVo cells, we tested sphere formation in vitro. As demonstrated in Figure 2, CD133+ LoVo cells had significantly increased number of sphere formation (CD133+ vs. CD133–: mean growth, 117.1% vs. 59.97%; difference, 57.12%; 95% CI, 50.78% to 63.46%; P<0.0001). Upon depletion of growth factors and exposed to 20% serum-containing medium, spheroid forming cells differentiated and became adherent (data not shown). In line with previous studies, tumor cells formed a typical flat monolayer with epithelioid morphology when sphere cells were differentiated and became adherent. Surface markers for cancer stem cells including nestin, musashi-1 were confirmed in colon tumor spheres as previously described (data not shown). To evaluate the tumorigenicity of CD133+ in animal xenograft model, we implanted FACS-sorted CD133+ LoVo cells (N=12) and CD133– LoVo cells (N=12). As demonstrated in Figure 3A, CD133+ LoVo cells were significantly tumorigenic when compared with CD133– LoVo cells (for tumors, CD133+ cells vs. CD133– cells: mean tumor volume on day 24, 90.42 vs. 11.85 cm3; mean difference, 78.57 cm3; 95% CI, 571.3 to 1,000; P≤0.0006).
Patient characteristics
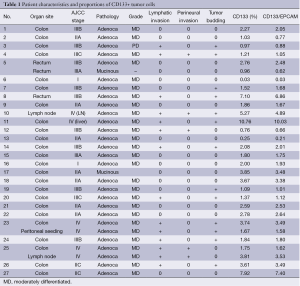
Based on the cell line and in vivo experiments, we next tested the proportion of CD133+ cells in tumor specimens and performed correlative analyses with clinicopathologic features. Between March 2010 and June 2010, 29 fresh tissue specimens were collected from 27 patients with colorectal cancer who underwent curative or palliative surgery at Samsung Medical Center. Mechanical and enzymatic dissociation into single cells was successful in all 29 tissue specimens, including 26 primary colorectal tumors, 2 lymph nodes, and 1 peritoneal seeding mass (Table 1). Histologically, 26 patients had adenocarcinomas and 1 had a mucinous adenocarcinoma. Twenty (74.1%) patients underwent low anterior or anterior resection, 6 underwent right hemicolectomy, and 1, with hereditary nonpolyposis colorectal cancer, underwent a subtotal colectomy for multifocal colon cancer. Ten of the 27 patients (37.0%) received postoperative chemotherapy or chemoradiation.
Full table
Single cell dissociated cells using surgically resected colorectal tumor specimens
Single cells dissociated from freshly extirpated colorectal cancer tissues were successfully obtained in all 29 specimens. All 29 specimens yielded at least >1×107 tumor cells with >70% viability after one freeze-thaw cycle. The dissociated cells were sorted according to the expression of CD133, a phylogenetically conserved cell surface marker associated with stem cells in glioblastomas, colon, and pancreatic cancers and osteosarcomas (12,13). Of the 29 tissue samples, 8 (27.6%) contained >3% CD133+ tumor cells. To confirm their epithelial origin, we sorted cells using antibody to EpCAM. We found that >90% of the CD133+ cells were also EpCAM(+) (Table 1).
CD133+ tumor cells as tumorigenic cells
To assess the tumorigenicity of CD133+ tumor cells isolated from colorectal tumor specimens, we tested their ability to form spheres in vitro. CD133+ tumor cells formed significantly more spheres than CD133– tumor cells (Figure 3A,B), with samples containing >3% CD133+ cells significantly more likely to form spheres than samples containing ≤3% CD133+ cells [36.4% (4/11) vs. 0% (0/16); P=0.019] (Table 2). Upon depletion of growth factors and >48-h exposure to medium containing 20% serum, these spheroid forming cells differentiated into tumor cells and became adherent (Figure 3B). In addition, the colon tumor spheres expressed surface markers for cancer stem cells, including nestin and musashi-1 (data not shown).

Full table
Correlation among sphere formation, high CD133+ percentage, and clinicopathologic features
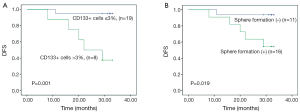
We also assessed the correlation between sphere formation and several clinicopathologic factors known to be associated with poor prognosis in patients with colorectal cancer, including tumor stage, lymphatic invasion, perineural invasion, and degree of differentiation. We found that sphere formation was not correlated with advanced stage (P=0.484) or perineural invasion (P=0.683), but was correlated with lymphatic invasion [54.5% (6/11) vs. 12.5% (2/16); P=0.033] and the presence of >3% CD133+ cells in tumor specimens [36.4% (4/11) vs. 0% (0/16); P=0.019] (Table 2). Moreover, we found that the presence of >3% CD133+ cells in tumor specimens was significantly correlated with advanced tumor stage (P=0.0044) (Table 3). Colorectal cancer patients with tumors containing >3% CD133+ tumor cells had a significantly shorter median DFS after surgery (22 months; 95% CI, 9.5-34.5 months) than did patients with tumors containing ≤3% CD133+ cells (median DFS not reached, P=0.001, Figure 4A). In addition, high spheroid formation was associated with significantly shorter DFS than the absence of spheroid formation (P=0.019, Figure 4B).

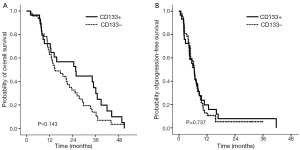
We also assessed whether CD133 positivity could predict survival in a cohort of 76 patients with metastatic colorectal cancer who had been treated with capecitabine plus oxaliplatin or S-1 plus oxaliplatin in a phase III trial (ClinicalTrials.gov, number NCT00677443). Immunohistochemical CD133 positivity correlated with the presence of >3% CD133+ tumor cells in tumors (data not shown). We found that CD133 was strongly positive in the luminal portion of tumor cells (Figure 3). Of the 76 patients, 29 (38.2%) showed strong immunopositivity for CD133. The overall response rate, defined as complete response plus partial response, was higher in patients immunohistochemically negative for CD133 than in than in those positive for CD133 (61.0% vs. 39.0%), although the difference was not statistically significant (P=0.528). And there were no significant difference regarding to OS and PFS between two groups (P=0.143 and P=0.737, Figure 5). The statistical significance of CD133+ positivity was not retained at multivariate level, however.
Discussion
Cancer stem cells are more resistant to standard cytotoxic chemotherapy (14) and/or radiotherapy (1,15) than non-stem cells. Drug resistance in cancer stem cells may be because of the increased expression of ATP-binding cassette drug transporters, such as ABCG2 and ABCB5, leading to the extrusion of chemotherapy agents from tumor cells (14,16,17). We hypothesized that patients with metastatic colorectal tumors containing >3% CD133+ cells would have higher staged and more invasive tumors, and therefore poorer survival outcomes, than patients with tumors containing ≤3% CD133+ cells. Most previous studies have focused on the in vitro tumorigenicity of CD133+ colon cancer cells, with few reporting on the clinical impact of tumors with >3-5% CD133+ cells on treatment outcome in patients with colorectal cancer. Since the feasibility of using of CD133+ cell lines to screen for inhibitors of stem cells or stem cell pathways in vitro is unclear, screening of CD133+ cells isolated directly from surgical specimens may be optimal.
Although it remains unclear whether CD133 is a robust marker of colon cancer stem cells (11), it is the most widely studied marker in colorectal cancer. Although other markers, such as CD44 (18) and CD166 (19) have been assessed as cancer stem cell markers, the combination of each with CD133 was no more specific than CD133 alone. It has been shown that trypsin could alter cell proteome through enzymatically cleaving of some cell surface markers (20). Tabatabaei et al. recently reported that trypsin used for enzymatic digestion affected both the percentage and expression of HLA-I and CD105 in human amniotic epithelial cells (21). However, established CRC cell lines routinely grown as adherent monolayers under serum-containing conditions showed a highly variable CD133 surface expression (22). We also used trypsin-EDTA to detach LoVo cells. Although it might affect the level of CD 133 expression in LoVo cell while sorting by flow cytometry using a FACS, the result that CD133+ LoVo cells were significantly associated in colony formation and sphere formation was same as we expected. The proportion of CD133+ cells in our tumor samples ranged from 0.03% to 10.76%. We successfully tested spheroid formation by single cells isolated from 27 of the 29 tumor specimens; we found that cells from 11 samples formed spheres in serum-free medium. We observed a strong correlation between sphere formation and lymphatic invasion (P=0.033), a factor significantly associated with poor prognosis in patients with colorectal cancer. In line with our results, the positive correlation between the percentage of CD133+ cells in CRC patient tumors and the presence of LN metastasis was recently reported (23). We also found that the presence in tumor specimens of >3% CD133+ cells was significantly associated with sphere formation (P=0.019), suggesting that CD133 is a valid marker for spheroid formation by colorectal cancer cells. This could be explained with the microenvironment difference between CD133+ and CD133– cells that genes regarding ECM proteins are downregulated in CD133+ cell while genes involved in degradation of ECM proteins, signal transduction and transcription for cell surface receptors CD133+ are upregulated (24).
In addition, we found that patients with tumors containing >3% CD133+ cells had a significantly shortened DFS than patients with tumors containing ≤3% CD133+ cells (P=0.001). Similarly, cells resistant to both 5-fluorouracil and oxaliplatin were significantly enriched in the expression of CD133 and CD44 (25). In our in vitro experiment, CD133+ LoVo cells were more resistant to 5-FU when compared with CD133– LoVo cells (Figure 1B, 1 μg/mL 5-FU CD133+ vs. 1 μg/mL 5-FU CD133–: mean growth, 51.23% vs. 14.09%; difference, 37.14%; 95% CI, 33.55% to 40.74%; P<0.0001). In another studies, high levels of CD133 mRNA strongly predicted short relapse-free survival and OS in colorectal cancer patients (12) and enrichment of CD133+ cancer cells during preoperative RCT in rectal cancer patients was correlated with minor local tumor response, increased distant cancer recurrence, and decreased survival (26). A recent meta-analysis study showed a significant correlation between CD133 expression and 5-year OS rate as well as DFS rate in CRC patients (27). Nevertheless, the correlation between the percentage of CD133-positive tumor cells and treatment outcome should be validated in a larger number of patients. Currently, we are performing gene expression profiling of CD133+ tumor cells isolated from patients with primary colorectal cancer and synchronous metastatic liver cancer.
Because of technical difficulties in sorting and isolating CD133+ tumor cells from surgical specimens using FACs, we used immunohistochemistry to test the prognostic ability of CD133 positivity in a different patient cohort. We recently reported the results of a phase clinical III trial comparing TS-1/oxaliplatin with capecitabine/oxaliplatin as first-line chemotherapy in patients with metastatic colorectal cancer (28). We tested 76 patients enrolled in that clinical trial whose tissue was available for CD133 immunohistochemical stain and found that >15% of tumor cells were positive for CD133 in 29 (38%) patients. These patients tended to be less responsive to chemotherapy than patients with CD133-negative tumors (P=0.528). Although there was no statistically significant difference in OS and PFS between patients with CD133 positive and CD133 negative in metastatic disease (Figure 5), the CD133 positivity in IHC was predictive of poor prognosis in colorectal cancer after surgery with statistical significance (Figure 4A), further experiments will be needed to confirm if CD133 positivity correlates with the actual proportion of CD133+ cells in single cell suspensions derived from surgical specimens. Furthermore, it will be needed to evaluate CD133 expression in circulating tumor cells from colorectal cancer for detecting CD133 in situ along with the immunochemistry and RT-PCR method. The clinical impact of CD133 positivity on the prediction of patients’ outcome and treatment practice for colorectal cancer should be elucidated.
Conclusions
In conclusion, we have shown that CD133 is a valid marker to identify cancer stem cells in fresh, surgically resected colorectal cancer samples. We found that 28% of these tumor specimens contained >3% CD133+ cancer stem cells. Using this living tumor bank and the isolated CD133+ tumor cells, we plan to screen for novel stem cell inhibitors.
Acknowledgements
This study was supported by a grant from the Samsung Biomedical Research Institute (#SBRI-CB11092, CB0220).
Disclosure: The authors declare no conflict of interest.
References
- Yeung TM, Mortensen NJ. Colorectal cancer stem cells. Dis Colon Rectum 2009;52:1788-96. [PubMed]
- Dylla SJ, Beviglia L, Park IK, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One 2008;3:e2428. [PubMed]
- Miraglia S, Godfrey W, Yin AH, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 1997;90:5013-21. [PubMed]
- Uchida N, Buck DW, He D, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A 2000;97:14720-5. [PubMed]
- Lee A, Kessler JD, Read TA, et al. Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci 2005;8:723-9. [PubMed]
- Singh SK, Clarke ID, Hide T, et al. Cancer stem cells in nervous system tumors. Oncogene 2004;23:7267-73. [PubMed]
- Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007;132:2542-56. [PubMed]
- Oshima Y, Suzuki A, Kawashimo K, et al. Isolation of mouse pancreatic ductal progenitor cells expressing CD133 and c-Met by flow cytometric cell sorting. Gastroenterology 2007;132:720-32. [PubMed]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111-5. [PubMed]
- O’Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106-10. [PubMed]
- Shmelkov SV, Butler JM, Hooper AT, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133– metastatic colon cancer cells initiate tumors. J Clin Invest 2008;118:2111-20. [PubMed]
- Artells R, Moreno I, Díaz T, et al. Tumour CD133 mRNA expression and clinical outcome in surgically resected colorectal cancer patients. Eur J Cancer 2010;46:642-9. [PubMed]
- Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med 2007;58:267-84. [PubMed]
- Lou H, Dean M. Targeted therapy for cancer stem cells: the patched pathway and ABC transporters. Oncogene 2007;26:1357-60. [PubMed]
- Rich JN. Cancer stem cells in radiation resistance. Cancer Res 2007;67:8980-4. [PubMed]
- Hirschmann-Jax C, Foster AE, Wulf GG, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A 2004;101:14228-33. [PubMed]
- Schatton T, Frank MH. Cancer stem cells and human malignant melanoma. Pigment Cell Melanoma Res 2008;21:39-55. [PubMed]
- Du L, Wang H, He L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res 2008;14:6751-60. [PubMed]
- Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 2007;104:10158-63. [PubMed]
- Huang HL, Hsing HW, Lai TC, et al. Trypsin-induced proteome alteration during cell subculture in mammalian cells. J Biomed Sci 2010;17:36. [PubMed]
- Tabatabaei M, Mosaffa N, Nikoo S, et al. Isolation and partial characterization of human amniotic epithelial cells: the effect of trypsin. Avicenna J Med Biotechnol 2014;6:10-20. [PubMed]
- Dittfeld C, Dietrich A, Peickert S, et al. CD133 expression is not selective for tumor-initiating or radioresistant cell populations in the CRC cell line HCT-116. Radiother Oncol 2010;94:375-83. [PubMed]
- Silinsky J, Grimes C, Driscoll T, et al. CD 133+ and CXCR4+ colon cancer cells as a marker for lymph node metastasis. J Surg Res 2013;185:113-8. [PubMed]
- Chao C, Carmical JR, Ives KL, et al. CD133+ colon cancer cells are more interactive with the tumor microenvironment than CD133– cells. Lab Invest 2012;92:420-36. [PubMed]
- Dallas NA, Xia L, Fan F, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res 2009;69:1951-7. [PubMed]
- Sprenger T, Conradi LC, Beissbarth T, et al. Enrichment of CD133– expressing cells in rectal cancers treated with preoperative radiochemotherapy is an independent marker for metastasis and survival. Cancer 2013;119:26-35. [PubMed]
- Chen S, Song X, Chen Z, et al. CD133 expression and the prognosis of colorectal cancer: a systematic review and meta-analysis. PLoS One 2013;8:e56380. [PubMed]
- Hong YS, Lee J, Kim KP, et al. Multicenter phase II study of second-line bevacizumab plus doublet combination chemotherapy in patients with metastatic colorectal cancer progressed after upfront bevacizumab plus doublet combination chemotherapy. Invest New Drugs 2013;31:183-91. [PubMed]


