
Limited bias effect of intratumoral heterogeneity on genetic profiling of hepatocellular carcinoma
Introduction
Utilization of next-generation sequencing (NGS) to profile tumor sample or biopsy for genetic variations potentially indicative of target drugs and then prescribe matched agents is increasingly accepted. Such attempt may find new sensitivity markers for established regimens or provide evidence supporting the off-label use of target drugs in unapproved diseases, thus facilitating the expansion of drug indications and the implement of precision medication. The precision medicine clinical trials—“umbrella” and “basket” studies are to evaluate the feasibility of such genotype-directed therapy (1). Two target drugs, sorafenib and lenvatinib, have been approved as the first-line therapy for hepatocellular carcinoma (HCC) but the response rate and treatment effect of sorafenib are not satisfactory (2) and large-scale clinical application of lenvatinib is just ongoing; novel agents are still needed. More important, neither of them have been reported of specific drug sensitivity/resistance markers. Thus, sequencing HCC to find genetic variations predicting drug response or suggesting the use of other target drugs with/without indications in HCC is rather promising (3).
Yet, the innate heterogeneous nature of cancer hinders accurate genome profiling: traditional single-site tumor sample or biopsy could hardly represent the full landscape of HCC genome due to intratumoral heterogeneity (ITH) and while such bias may be subtle, it could eventually lead to drug resistance and treatment failure (4). A thorough knowledge of the influence posed by ITH on genotyping would be helpful. Previously, we and other researchers have reported ITH of HCCs; nevertheless, how and to what extent would ITH impede precise tumor genotyping remain unknown. Although prospective studies involving multi-center and large-scale cases, such as what have been done in clear-cell renal cell carcinoma (5), might adequately interpret, it needs exquisite design and could be time-consuming and fund-costing. Herein, we reviewed recent published data on ITH of HCC and extracted the sequencing information to make data reanalysis in order to preliminarily clarify whether genetic information profiled from single tumor sample would be biased by ITH.
Materials and methods
Definition of actionable genes
If the protein product of a genomic aberration could be potentially targeted by a specific targeted drug, this mutant gene was claimed to be actionable. The precision medicine trails were used as reference to identify potential actionable genes in HCC. Genes reported actionable or relevant with target therapy response in HCC by former studies were also included. Candidate genes with target therapy potential were screened and crosschecked by two investigators independently.
Literature retrieval and data analysis
We used the following key items to search studies: intratumoral heterogeneity; intratumor heterogeneity; ITH; tumor heterogeneity; liver cancer; hepatocellular carcinoma; HCC. Clinicopathological parameters and sequencing data were acquired. The variations were classified according to our previously reported standards as well as the measurement of ITH extent. Spearman’s correlation analysis was used to evaluate the correlation between ITH extent and sequencing parameters. For each analysis, the correlation efficient r and P value were calculated. P<0.05 was considered to be statistically significant. All analyses were conducted with SPSS 18. This study was approved by the Institutional Review Board of Zhongshan Hospital, Fudan University. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Results
Identification of genes relevant with target therapy
As previously reported, genetic aberrations which could be targeted either directly or indirectly by Food & Drug Administration (FDA) approved or investigational agents undergoing clinical trials are considered potentially druggable (6). Mutants leading to target therapy resistance are also regarded as actionable. Next we thoroughly checked the FDA approved target drugs for oncology as well as the precision medicine trials including Targeted Agent and Profiling Utilization Registry (TAPUR) (7) and National Cancer Institute (NCI) initiated Molecular Analysis for Therapy Choice (MATCH) and Molecular Profiling-based Assignment of Cancer Therapy (MPACT) (8). Recent trials which performed genome-driven target therapy or studies with large-scale genomic testing to screen actionable mutations were also consulted (9-17). Since we focused solely on the influence of ITH on genotyping, only genetic biomarkers were pooled as reference. Genetic signatures believed to be associated with the treatment response of sorafenib (18-24) were given special attentions. Totally, 72 genes were identified to be associated with treatment response of target drugs (ABL, AKT, ALK, ATM, ATR, BRAF, BRCA1/2, BTK, CCND1/2/3, CDK4/6, CDKN2A, cKIT, CSF1R, DDR2, EGFR, EPHA2, ERCC1, FBXW7, FGF3/4, FGF19, FGFR1/2/3, FLT3, FYN, GNAQ, GNA11, HER2, HRAS, IDH2, JAK1, KRAS, LCK, MET, MEK1/2, MLH1, MSH2, mTOR, NBN, NF1/2, NRAS, NTRK1/2/3, PARP1/2, PDGFR, PIK3CA, PIGF, PTCH1, PTEN, ROS1, RAD51, RANKL, RET, ROS1, SLC15A2, SLC22A1, SRC, TSC1/2, VEGFA, VEGFR1/2/3).
Literature search and inclusion
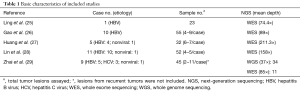
Literature retrieval identified five studies concerning the genetic ITH of HCC (25-29) (Table 1). Friemel et al. focused mainly on the morphological ITH of HCC by measuring the expression of the liver cell markers (30) and Buczak et al. used proteomics (31). The work by Xue et al. was performed in multiple intrahepatic lesions rather than in multiple regions within single lesion (32); thus the heterogeneity was intertumoral rather than intratumoral. Shi et al. reported the spatial and temporal heterogeneity of a patient with multifocal liver cancer (33), as well as the work by Xu et al. (34). Cases presented by Wang et al. were liver cancers with clinicopathological features of both HCC and intrahepatic cholangiocarcinoma (ICC) (35) while Dong et al. reported the ITH of ICC (36). Zheng et al. (37) and Duan et al. (38) studied heterogeneity of HCC at single-cell level. Li et al. reported the ITH of somatic mitochondrial DNA in HCC (39). These nine studies were excluded as well as former studies without genetic sequencing data (40-42). Of the included studies, the sequencing results were all available and we thoroughly reviewed the sequencing information of each tumor and classified the nonsynonymous genetic variations and copy number variations (if there were any) identified in each HCC as ubiquitous (present in all regions), private (present in single region) and heterogeneous (the left) as previously reported (27). Clinicopathological information of enrolled patients and their matched tumors in each study were also checked.
Full table
ITH extent and its influence on genetic variation profiling of HCC
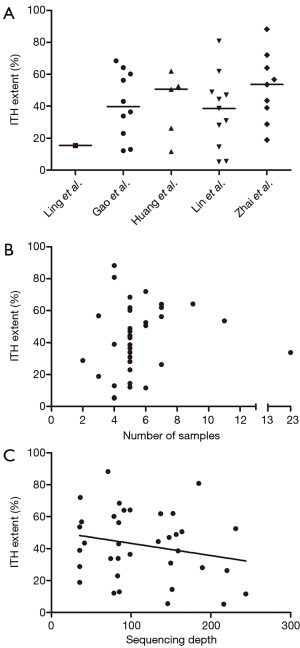
Totally, 207 tumor samples from 36 HCCs were assayed in the above studies (Table 1). Most patients (30/36) had hepatitis B virus (HBV) infection history, three patients were hepatitis C virus (HCV) infected and there were of nonviral background. Whole exome sequencing (WES) was predominantly used for genetic profiling (84.8% of all tumor samples) at mean sequencing depth ranging from 85×–211.3×. Upon classifying the genetic variations in each study using predefined standard, we found the ITH extent distributed in wide range from 5.21% to 88.27% (Figure 1A) and the distribution pattern was quite similar among studies (except the study with only one case). We then analyzed whether sampling strategy or sequencing condition might affect ITH extent. Neither the number of samples (Figure 1B; Spearman r=0.2642; P=0.12) nor sequencing depth (Figure 1C; Spearman r=–0.1571; P=0.36) had any significant correlations with ITH extent. Furthermore, no relations were found between tumor clinicopathological parameters [alpha-fetoprotein (AFP), gender, age, etiology, tumor-node-metastasis (TNM) stage, tumor size] and ITH extent.
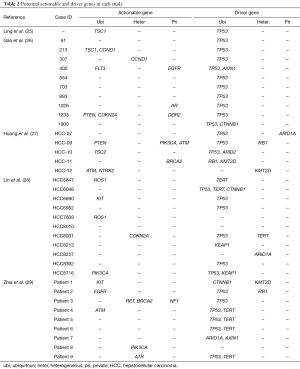
We then focused on the potential challenges posed by ITH on therapeutic targets profiling. The actionable genes in each study were mined out (Table 2). Totally, 30 therapeutic targets located in 18 genes were identified in 21 cases and 58.3% samples (21/36) were identified to harbor at least 1 therapeutic targets. Of the 30 variations, 17 were ubiquitous, 6 were heterogeneous, and 7 were private ones (Figure 2A). Since heterogeneous and private variations could not always be detected in single-site sampling strategy, the patients harboring ubiquitous variations stood as the undoubted ones with target therapy potential, under which circumstance, only 38.9% HCCs (14/36) were found to be drugable. Interestingly, this ratio was quite similar to the rate we found in real-world sequencing practice (38.6%) (27). Meanwhile, 19.4% HCCs (7/36) containing only heterogeneous and private variations may thus be biased by ITH.
Full table
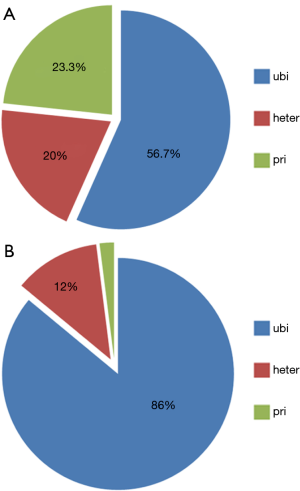
To further evaluate the impact of ITH on genetic profiling, we specially analyzed the heterogeneity of HCC driver genes in these cases. In case of genes with target therapy potential as well as tumor-driver ability, they were categorized as drugable ones (PTEN, TSC2, ATM, CDKN2A, and CCND1). Studies reporting and systematical review of NGS results of HCC, and online HCC sequencing databases were thoroughly checked and 15 highly frequently mutant genes were considered as candidate driver genes: TERT, TP53, CTNNB1, ARID1A, ARID2, AXIN1, RB1, CCNE1, KMT2D, RP6SKA3, IL6ST, NFE2L2, MLL4, KEAP1, APC. In total, we identified 50 genetic variations in 9 driver genes (TP53, TERT, CTNNB1, ARID1A, ARID2, AXIN1, KEAP1, KMT2D, RB1) and 91.7% (33/36) of the cases contained at least one driver gene (Table 2). Consistent with the fact that most HCCs in the studies were HBV-associated, TP53 demonstrated to be the most frequent driver gene (26/36) and unexpectedly, it was ubiquitously present in all these cases. Moreover, we found 86% (43/50) of the driver genes were ubiquitous ones, 6 (12%) were heterogeneous ones, and only one driver gene was privately existed (Figure 2B). This finding coincided with the result by Torrecilla et al. (43), which reported that truncal mutational events in HCC were ubiquitous across different regions of the same tumor in more than 85% cases—in other words, driver genes were less likely to be biased by ITH. Thus, single-site sampling could be enough to capture most, if not all, driver mutations in HCC.
Discussion
The ITH of HCC has been long recognized and former studies have used different strategies including immunohistochemistry, proteomics, DNA ploidy analysis, sequencing and even fluorodeoxyglucose (FDG) uptake (44) to measure the ITH extent of HCC. Recently, NGS-based genomic profiling of multi-regional samples have been widely used to evaluate ITH. However, the limited case analyzed in each study made it difficult to clearly and thoroughly elucidate how ITH might challenge accurate genomic profiling of HCC. What’s more, no previous work had gone a step further to particularly discuss the potential influence of ITH in target therapy, especially in the current era of precision medicine. Herein, we tried to answer the abovementioned questions by analyzing excised data: we made a thorough review and datamined relevant publications to make re-analysis in order to clarify whether and how ITH might influence precise genomic profiling.
An overview of the studies found ITH differed significantly among HCC cases and no associations between ITH and clinicopathological factors were observed. However, this should be cautiously taken since cases from different cohort may lead to bias, small sample size failed generate any significant association, and it’s unknown whether etiological and epidemiological factors could exert influence on ITH, although they closely interact with tumorigenesis and tumor biology. Intuitively, the more tumors are heterogeneous, the more their biological behaviors are malignant; but data shortage made it impossible to analyze ITH extent with tumor aggressiveness, such as differentiation, proliferation and vascular invasion. In other cancers like renal cell carcinoma (5,45) and non-small-cell lung cancer (46), tumor heterogeneity and evolution trajectories predict recurrence and prognosis, serving both intervention and surveillance. Moreover, we found that although many HCCs in the included datasets had TP53 as driver gene, they harboured different actionable genes, which may result from the heterogeneous tumor evolution process after tumorigenesis of HCC even under the same etiology (mainly HBV) and tumor driver gene (47). This necessitates the use of NGS or other assays to screen for potential therapeutic target before precision targeted therapy. Thus, future experimental studies addressing the interaction between ITH and tumor biology are needed and may bring new sights into the management of HCCs with high heterogeneity.
Although genotype-driven target therapy is widely advocated, few chances are provided for HCC patients. Neither sorafenib nor newly approved lenvatinib and regorafenib have clear-cut genetic indications. We thus referred to other cancer types to identify genes that could be potentially targeted in HCC. Oncogenes mediating tumor progression, signaling pathways interfering with target reagents, and genes with indications in other cancers were pooled. Despite by the fact that the percentage of HCCs with target therapy potential was similar to rate we found in clinical practice, nearly 20% HCCs with actionable targets might be left out due to ITH. However, this does not mean multi-site sampling should be employed to comprehensively identify targetable patients since the risk of repeated tumor biopsy and the cost-effectiveness of additional sample assays need to be concerned. Moreover, it has been reported that liquid biopsy such as circulating tumor DNA (ctDNA) could complement tissue biopsy to draw a full landscape of tumor genome and the application of ctDNA as surrogate to overcome tumor heterogeneity is increasingly recognized (48).
Surprisingly, we found the data re-analysis results coincided with the work by Torrecilla et al. (43). It’s generally accepted that driver/truncal mutations arise at early stages of cancer and are shared by nearly all malignant cells within the tumor. Torrecilla et al. characterized the early stages of hepatocarcinogenesis, namely dysplastic nodules and small lesions and found that mutations in TERT, TP53 and CTNNB1 were the most frequent and more than 85% of such driver mutations were also shared in more advanced lesions between different regions of the same tumor. This experimental evidence supports our finding that 85% driver genes were ubiquitously existed. Thus, single-site biopsy is capable to capture truncal events and under such circumstance, molecular classification based on truncal genes cluster could accurate reflect tumor biology of HCC.
Conclusions
This study systematically reviewed NGS studies concerning the genomic ITH of HCC and re-analyzed the sequencing data to thoroughly evaluate the extent of ITH and its impact on HCC tumor genome profiling. ITH varied among HCCs and it indeed hinders comprehensive and precise genomic profiling; however, the influences of ITH on identification of actionable and driver mutations seems to be limited since most genes of these kind were ubiquitously present in tumors. The routinely used single-site biopsy/sampling, when combined with targeted deep sequencing, may be enough for the clinical management of HCC and of high cost-effectiveness. Still, clinical evidence from real world is urgently needed to support our viewpoints on ITH and genome-driven target therapy of HCC.
Acknowledgments
Funding: This study was jointly funded by the National Natural Science Foundation of China (No. 81830102, 81802991, 81572823, 81772578) and STCSM (18YF1403600).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Renfro LA, An MW, Mandrekar SJ. Precision oncology: a new era of cancer clinical trials. Cancer Lett 2017;387:121-26. [Crossref] [PubMed]
- Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology 2016;150:835-53. [Crossref] [PubMed]
- Chan SL, Wong AM, Lee K, et al. Personalized therapy for hepatocellular carcinoma: where are we now? Cancer Treat Rev 2016;45:77-86. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613-28. [Crossref] [PubMed]
- Turajlic S, Xu H, Litchfield K, et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 2018;173:595-610.e11. [Crossref] [PubMed]
- Meric-Bernstam F, Johnson A, Holla V, et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst 2015. [Crossref] [PubMed]
- Brower V. American society of clinical oncology developing first clinical trial. J Natl Cancer Inst 2015;107:djv356. [Crossref] [PubMed]
- Do K, O'Sullivan Coyne G, Chen AP. An overview of the NCI precision medicine trials-NCI MATCH and MPACT. Chin Clin Oncol 2015;4:31. [PubMed]
- Millis SZ, Ikeda S, Reddy S, et al. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19784 diverse solid tumors. JAMA Oncol 2016;2:1565-73. [Crossref] [PubMed]
- Kaplan R, Maughan T, Crook A, et al. Evaluating many treatments and biomarkers in oncology: a new design. J Clin Oncol 2013;31:4562-8. [Crossref] [PubMed]
- Le Tourneau C, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324-34. [Crossref] [PubMed]
- Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol 2015;33:2753-62. [Crossref] [PubMed]
- Beltran H, Eng K, Mosquera JM, et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 2015;1:466-74. [Crossref] [PubMed]
- Stockley TL, Oza AM, Berman HK, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med 2016;8:109. [Crossref] [PubMed]
- Tanabe Y, Ichikawa H, Kohno T, et al. Comprehensive screening of target molecules by next-generation sequencing in patients with malignant solid tumors: guiding entry into phase I clinical trials. Mol Cancer 2016;15:73. [Crossref] [PubMed]
- Cousin S, Grellety T, Toulmonde M, et al. Clinical impact of extensive molecular profiling in advanced cancer patients. J Hematol Oncol 2017;10:45. [Crossref] [PubMed]
- Schwaederle M, Chattopadhyay R, Kato S, et al. Genomic alterations in circulating tumor DNA from diverse cancer patients identified by next-generation sequencing. Cancer Research 2017;77:5419-27. [Crossref] [PubMed]
- Herraez E, Lozano E, Macias RI, et al. Expression of SLC22A1 variants may affect the response of hepatocellular carcinoma and cholangiocarcinoma to sorafenib. Hepatology 2013;58:1065-73. [Crossref] [PubMed]
- Sawey ET, Chanrion M, Cai C, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell 2011;19:347-58. [Crossref] [PubMed]
- Arao T, Ueshima K, Matsumoto K, et al. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology 2013;57:1407-15. [Crossref] [PubMed]
- Scartozzi M, Faloppi L, Svegliati Baroni G, et al. VEGF and VEGFR genotyping in the prediction of clinical outcome for HCC patients receiving sorafenib: the ALICE-1 study. Int J Cancer 2014;135:1247-56. [Crossref] [PubMed]
- Horwitz E, Stein I, Andreozzi M, et al. Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov 2014;4:730-43. [Crossref] [PubMed]
- Lee YS, Kim BH, Kim BC, et al. SLC15A2 genomic variation is associated with the extraordinary response of sorafenib treatment: whole-genome analysis in patients with hepatocellular carcinoma. Oncotarget 2015;6:16449-60. [PubMed]
- Kaibori M, Sakai K, Ishizaki M, et al. Increased FGF19 copy number is frequently detected in hepatocellular carcinoma with a complete response after sorafenib treatment. Oncotarget 2016;7:49091-98. [Crossref] [PubMed]
- Ling S, Hu Z, Yang Z, et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci U S A 2015;112:E6496-505. [Crossref] [PubMed]
- Gao Q, Wang ZC, Duan M, et al. Cell culture system for analysis of genetic heterogeneity within hepatocellular carcinomas and response to pharmacologic agents. Gastroenterology 2017;152:232-42.e4. [Crossref] [PubMed]
- Huang A, Zhao X, Yang XR, et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J Hepatol 2017;67:293-301. [Crossref] [PubMed]
- Lin DC, Mayakonda A, Dinh HQ, et al. Genomic and epigenomic heterogeneity of hepatocellular carcinoma. Cancer Res 2017;77:2255-65. [Crossref] [PubMed]
- Zhai W, Lim TK, Zhang T, et al. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat Commun 2017;8:4565. [Crossref] [PubMed]
- Friemel J, Rechsteiner M, Frick L, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res 2015;21:1951-61. [Crossref] [PubMed]
- Buczak K, Ori A, Kirkpatrick JM, et al. Spatial tissue proteomics quantifies inter- and intratumor heterogeneity in hepatocellular carcinoma (HCC). Mol Cell Proteomics 2018;17:810-25. [Crossref] [PubMed]
- Xue R, Li R, Guo H, et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 2016;150:998-1008. [Crossref] [PubMed]
- Shi JY, Xing Q, Duan M, et al. Inferring the progression of multifocal liver cancer from spatial and temporal genomic heterogeneity. Oncotarget 2016;7:2867-77. [PubMed]
- Xu LX, He MH, Dai ZH, et al. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol 2019. [Epub ahead of print]. [PubMed]
- Wang A, Wu L, Lin J, et al. Whole-exome sequencing reveals the origin and evolution of hepato-cholangiocarcinoma. Nat Commun 2018;9:894. [Crossref] [PubMed]
- Dong LQ, Shi Y, Ma LJ, et al. Spatial and temporal clonal evolution of intrahepatic cholangiocarcinoma. J Hepatol 2018;69:89-98. [Crossref] [PubMed]
- Zheng H, Pomyen Y, Hernandez MO, et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 2018;68:127-40. [Crossref] [PubMed]
- Duan M, Hao J, Cui S, et al. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res 2018;28:359-73. [Crossref] [PubMed]
- Li X, Guo X, Li D, et al. Multi-regional sequencing reveals intratumor heterogeneity and positive selection of somatic mtDNA mutations in hepatocellular carcinoma and colorectal cancer. Int J Cancer 2018;143:1143-52. [Crossref] [PubMed]
- Okada S, Ishii H, Nose H, et al. Intratumoral DNA heterogeneity of small hepatocellular carcinoma. Cancer 1995;75:444-50. [Crossref] [PubMed]
- Saeki R, Nagai H, Kaneko S, et al. Intratumoral genomic heterogeneity in human hepatocellular carcinoma detected by restriction landmark genomic scanning. J Hepatol 2000;33:99-105. [Crossref] [PubMed]
- An FQ, Matsuda M, Fujii H, et al. Tumor heterogeneity in small hepatocellular carcinoma: analysis of tumor cell proliferation, expression and mutation of p53 AND beta-catenin. Int J Cancer 2001;93:468-74. [Crossref] [PubMed]
- Torrecilla S, Sia D, Harrington AN, et al. Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J Hepatol 2017;67:1222-31. [Crossref] [PubMed]
- Dong A, Yu H, Wang Y, et al. FDG PET/CT and enhanced CT imaging of tumor heterogeneity in hepatocellular carcinoma: imaging-pathologic correlation. Clin Nucl Med 2014;39:808-10. [Crossref] [PubMed]
- Turajlic S, Xu H, Litchfield K, et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 2018;173:581-94.e12. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Davis A, Gao R, Navin N. Tumor evolution: linear, branching, neutral or punctuated? Biochim Biophys Acta Rev Cancer 2017;1867:151-61.
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]


