Gene mutations distinguishing gastric from colorectal and esophageal adenocarcinomas
Introduction
Literature and our previous work indicated that tumor stage is the only significant predictor for survival of gastric cancer. Specifically, patients with tumors of localized stage have 70% probability of 5-year survival while patients with tumors of loco-regional stage have 30% probability of 5-year survival and those with metastasized cancer have 5% probability of 5-year survival (1,2).
There are three classification systems which classify gastric tumors into distinct subgroups beyond tumor stage. The Lauren histological classification groups gastric tumors into two main types of cell histology—intestinal and diffuse (3). The WHO classification divides gastric tumors into papillary, tubular, mucinous, and poorly cohesive carcinomas (4). Recently, a genetic study published in the Journal of Nature introduces a new molecular classification of gastric tumors into four genetic subgroups: EBV-positive tumors, microsatellite unstable tumors, genomically stable tumors, and chromosomally unstable tumors (5). While numerous studies have linked diffuse histology and poorly differentiated tumors to poor prognosis, none of these classification systems explains the variability in patient survival better than tumor stage.
In our review of the literature, we found one genetic study that linked genetic mutations to patient survival. Performing genetic profiling of 521 gastric tumors of patients from 4 medical centers in different countries, it found a cluster of 171 genes associated with worse survival (6). Unfortunately, this finding has little clinical and translational utility because it is challenging to target hundreds of genes simultaneously via pharmacologic or genetic manipulation.
In search for genetic factors that could better explain survival beyond tumor stage, we focused on finding genes that are mutated in only gastric tumors, and once mutated, cause tumors to progress to advanced stages. However, we observed that most mutations found in gastric cancer are also found in colorectal and esophageal cancer. Therefore, the challenge remained to identify mutations that distinguish gastric from colorectal and esophageal cancers. We sought to identify mutations that accounted for the unique phenotypic features of gastric tumors using open-access cancer genomics data.
Methods
Ethics approval for this study was exempted by the Institutional Review Board at the University of California in Irvine because this study is a non-human subject research, making use of public datasets with non-identifiable subjects (7).
Data collection
We downloaded a total of 13 open-access cancer genomics datasets, including 7 gastric, 4 colorectal, and 2 esophageal adenocarcinoma datasets from cBioPortal website (8,9). These datasets were last accessed on May 8, 2019. Names of the studies, years from which these datasets were generated, and the numbers of subjects are listed in Table S1. The datasets included demographic, clinical, and genetic variables of which descriptions can be found at the NCI Genomic Data Commons website (10). The following variables were selected for the analysis: age, gender, race, pathologic stage, histology, Hugo symbol, chromosome, start nucleotide position, end nucleotide position, mutation type, mutation classification, nucleotide change (HGVSc), and amino acid change (HGVSp).

Full table
Genetic variables were linked to demographic and clinical variables by subject identifier and tumor sample identifier. Since there was only one primary tumor sample per subject, the sample identifier was used interchangeably as the subject identifier. Each gene was identified by Hugo symbol which is human gene nomenclature while a specific mutation of a gene was defined by Hugo symbol, the affected chromosome, and the lowest numeric nucleotide position of the reported mutation on the genomic reference sequence. The final dataset used for analysis included three-nested levels of data: gene-specific mutations which were nested within genes, which in turn were nested within samples.
Statistical analyses
To identify mutated genes discriminating diffuse histology from intestinal histology of gastric tumors, we compared the percentage of diffuse histology between subjects with and without a specific mutated gene using two-sample t-tests, adjusting for multiple gene testing. This analysis was applicable to only gastric cancer subjects with valid diffuse and intestinal histology.
To identify mutated genes distinguishing gastric from colorectal and esophageal tumors, we first quantified each subject’s pathologic TNM stage into a continuous score from 1 to 8 as follows: IA=1, IB=2, IIA=3, IIB=4, IIIA=5, IIIB=6, IIIC=7, and IV=8. Then we compared the mean stage among four groups: non-carriers, gastric mutation carriers, colorectal mutation carriers, and esophageal mutation carriers. Mutated genes were considered as significant if the following criteria were met. Statistical significance: difference in the mean stage among the four comparison groups must be significant beyond random chance based on the overall F test and the post-hoc two-sample t tests with P value <0.05 after adjusted for multiple gene testing using Benjamini-Hochberg’s false discovery rate (11). Equivalently, the unadjusted P values would be <10−6. Clinical significance: a mutated gene would be deemed as clinically significant if it discriminates the three tumor types into localized stages versus locoregional stages.
For the mutated genes that were found to be significantly associated with gastric histology and advanced tumor stages, we compared the genetic characteristics of these mutations against the background which included all genes in the datasets. Genetic characteristics included mutation type (i.e., single nucleotide polymorphism SNP, deletion DEL, insertion INS, etc.), mutation classification (i.e., missense, nonsense, frameshift, etc.), and single nucleotide substitution (i.e., A>C, A>G, A>T, etc.). To determine the impact of the genetic characteristics of the mutated genes upon protein functions, sequence convergence and functional impact were analyzed using permutation test and PolyPhen-2 score, respectively. PolyPhen-2 score is a probability value from 0 to 1, predicting the possible impact of amino acid substitutions on the stability and function of human proteins using structural and comparative evolutionary considerations (12).
Results
Demographic characteristics
After removing duplicated subjects across the datasets, we identified a total of 2,264 unique subjects. We excluded the subjects whose pathologic stages were missing, leaving a total of 1,915 subjects with valid pathologic stages, including 564 subjects with primary tumors of gastroesophageal adenocarcinoma, 1,140 subjects with primary tumors of colorectal adenocarcinoma, and 211 subjects with primary tumors of esophageal adenocarcinoma in the analysis. Table 1 presents descriptive statistics of the demographic, clinical, and genetic variables of the study subjects. Majority of the subjects were between 40–80 years old (99%) while very few subjects were younger than 40 years (1%). Mean age was 68 years (SD=10). Fifty four percent were female and 46% were male. The most common ethnicity was Caucasian (37%) while minorities were rare with high rates of missing ethnicity data (44%).
Full table
Clinical characteristics
Pathologic stages included 20% stage I, 35% stage II, 31% stage III, and 14% stage IV. By anatomical location, there were 30% gastric tumors, 59% colorectal tumors, and 11% esophageal tumors. The histology of the gastric tumors included 61% intestinal, 23% diffuse, 15% mixed, and 1% missing.
Genetic characteristics
We excluded the genetic mutations that were non-exon and silent because of their presumably non-functional effects on proteins, leaving about 25,000 genes and 690,000 non-silent exon mutations in the genetic analysis. Mutation types were 86% SNP, 11% DEL, and 3% INS. Mutation classification included 80% missense, 12% frameshift, 5% nonsense, 2% splice, and 1% in-frame. Single variant substitutions included 56% G>A, 16% G>T, 14% A>G, 7% A>C, 4% A>T, and 3% G>C. Although we observed that G>A was the predominant signature, it is unclear how this signature is related to the cancer etiology.
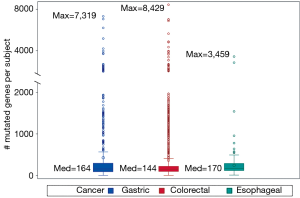
Genetic mutations were heterogeneous across subjects, varying from 8 to 8,429 mutated genes per subject. No two subjects shared the same genetic mutation profile. Figure 1 compares gene diversity among the three cancer types. Gastric and colorectal cancer subjects had the most diverse mutations with a maximum of 7,319 and 8,429 mutated genes per subject respectively while esophageal cancer subjects had significantly fewer mutated genes with a maximum of 3,459 (P=0.005).
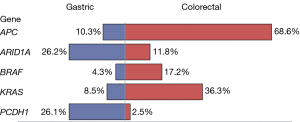
The most common mutated genes include TP53 (60%), TTN (54%), APC (45%), and KRAS (25%). Most mutated genes found in gastric tumors were also found in colorectal tumors. It was not the mutated gene itself but rather the frequency of a mutated gene that distinguished the two cancer types. For example, the mutated APC gene occurred in 10% of the gastric tumors as compared to 69% of the colorectal tumors; in contrast, the mutated ARID1A gene occurred in 26% of the gastric tumors as compared to 12% of the colorectal tumors. Figure 2 lists the top five mutated genes with contrasting frequencies, including ARID1A and PCDH1 which were more common in gastric tumors and APC, BRAF, KRAS which were more common in colorectal tumors.
Mutated genes associated with diffuse-gastric histology
We found two mutated genes discriminating diffuse histology from intestinal histology of gastric tumors: the E-cadherin CDH1 gene and the cell adhesion RHOA gene (Table 2). Subjects with these mutated genes were 5 to 6 times more likely to have diffuse histology then subjects without these mutated genes (odds ratios =5.7–6.6, P<10−6). Specifically, the percentage of diffuse histology was 64–70% among subjects with these mutated genes as compared to 24–26% among subjects without these mutated genes. In contrast, the percentage of intestinal histology was 21–31% among subjects with these mutated genes as compared to 64–65% among subjects without these mutated genes (odds ratio =0.1–0.2, P<10−6).

Full table
Mutated gene distinguishing gastric from colorectal and esophageal tumors
We found CDH1 to be the only mutated gene distinguishing gastric from colorectal and esophageal tumors: gastric cancer subjects who carried this mutated gene were more likely to have loco-regional tumors while colorectal and esophageal cancer subjects who carried this mutated gene were more likely to have localized tumors (Table 3). Specifically, the percentage of loco-regional stages was 66% among gastric mutation carriers as compared to 28% among colorectal mutation carriers and 0% among esophageal mutation carriers (P<10−6).

Full table
CDH1 and RHOA recurrent hotspots and functional impacts
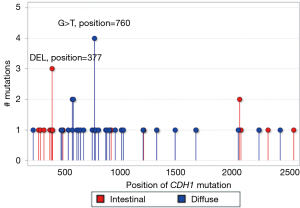
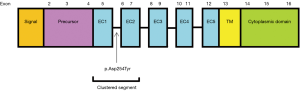
We defined a recurrent hotspot as the mutation of a gene that occurred at the same nucleotide position in the gene sequence in three or more tumors. Figure 3 displays the nucleotide positions of CDH1 recurrent hotspots in diffuse and intestinal tumors. In diffuse tumors, there was one recurrent hotspot involving G>T missense substitutions at nucleotide position 760 on exon number 6 in four tumors which resulted in protein change p.Asp254Tyr. This hotspot, affecting the calcium binding pocket connecting the extracellular cadherin domains EC1 and EC2, had the highest impact on protein function with PolyPhen-2 score of 1 (on a scale from 0 to 1). In intestinal tumors, there was one recurrent hotspot involving deletion mutations at nucleotide position 377 in three tumors. More importantly, 65% of CDH1 mutations in diffuse tumors clustered in the sequence segment between nucleotide position 500 and 1,000 on exons 5 to 7, which affect the extracellular cadherin domains EC1 and EC2 and the calcium binding pocket connecting these two domains (Figure 4). In contrast, the majority of CDH1 mutations in intestinal tumors were scattered in two opposite segments, before nucleotide position 500 and after nucleotide position 1,000. Permutation test indicated that the contrasting CDH1 nucleotide positions between diffuse and intestinal tumors were statistically significant (P<0.0001).
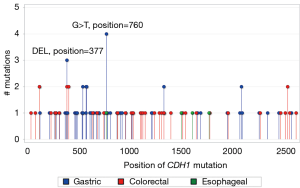
We compared the nucleotide positions of CDH1 mutations in gastric, colorectal, and esophageal tumors in the lollipop graph in Figure 5. There were two recurrent hotspots in gastric tumors (the same hotspots as mentioned in Figure 3) while there was no hotspot in colorectal and esophageal tumors. In addition, about 60% of CDH1 mutations in gastric tumors clustered in the sequence segment between nucleotide position 500 and 1,000 while CDH1 mutations in colorectal and esophageal tumors were scattered randomly. Permutation test indicated that the contrasting patterns of CDH1 nucleotide positions between gastric and colorectal/esophageal tumors were statistically significant (P<0.0001).
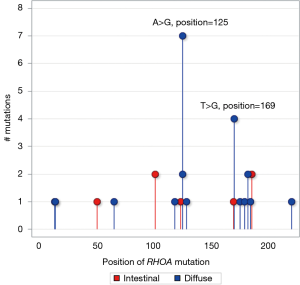
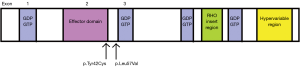
In Figure 6, we compared RHOA recurrent hotspots in diffuse and intestinal tumors. In diffuse tumors, there were two recurrent hotspots involving A>G missense substitutions at nucleotide position 125 on exon number 2 resulting in protein change p.Tyr42Cys in seven tumors, and T>G missense substitutions at nucleotide position 169 on exon number 3 resulting in protein change p.Leu57Val in four tumors. The protein change p.Tyr42Cys is located in the effector binding domain while the protein change p.Leu57Val is located at the border of the GDP/GTP binding domain (Figure 7). Both hotspots had high impact on protein function with PolyPhen-2 probability score of 0.718 and 0.999, respectively. There was no RHOA hotspot in intestinal tumors.
In Table S2, we listed detailed information of CDH1 and RHOA mutations, including anatomical location, histology, nucleotide change (HGVSc), amino acid change (HGVSp), mutation type, and mutation classification.
Full table
CDH1 and RHOA genetic landscapes against background
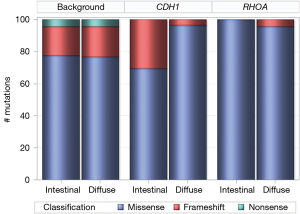
We compared the distribution of missense, nonsense, and frameshift mutations of CDH1 and RHOA between diffuse and intestinal gastric tumors, against the background which included mutations of all genes (Figure 8). The mutation types of the background were identical between diffuse and intestinal tumors with 78% missense, 18% frameshift, and 4% nonsense. In contrast to the background, the distribution of CDH1 mutation types involved 97% missense mutations in diffuse tumors versus 70% missense mutations in intestinal tumors (P=0.0054). Although different from the background, the distribution of RHOA mutation types was not statistically different between diffuse and intestinal tumors: 96% missense mutations in diffuse tumors versus 100% missense mutations in intestinal tumors (P=0.5830).
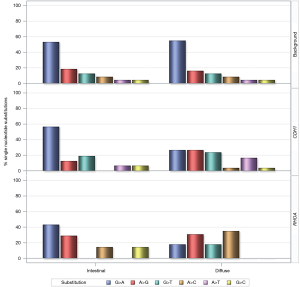
In addition, we compared the distribution of single nucleotide substitutions of CDH1 and RHOA missense mutations between diffuse and intestinal gastric tumors, against the background (Figure 9). Substitutions in the background were identical between diffuse and intestinal tumors with G>A as the most common missense substitution. In contrast to the background, G>A was significantly less common among CDH1 mutations and RHOA mutations in diffuse tumors than intestinal tumors. Similarly, the single nucleotide substitutions of CDH1 and RHOA missense mutations was different among gastric, colorectal, and esophageal tumors (data not shown). It is unclear how the dissimilarities in single nucleotide substitutions between histology types and cancer types are related to the cancer etiology.
Discussion
Our genetic analysis of 1,915 subjects with gastrointestinal malignancies showed approximately 25,000 mutated genes in the tumors. At the subject level, the number of mutated genes varied from 10 to 8,000 per subject; no two subjects shared the same mutation profile. Of the three cancer types, gastric and colorectal tumors had the most gene diversity with the maximum number of mutated genes up to 7,000–8,000 per subject while esophageal tumors had only 3,500 per subject at the maximum. This finding was consistent with a global cancer study which found gastric and colorectal cancers with the largest gene diversity while esophageal cancer with moderate gene diversity (13). At the gene level, most mutations found in gastric tumors were also found in colorectal tumors. Therefore, it was not the mutated gene itself but rather the frequency of a mutated gene that distinguished the two cancer types. The top five genes with contrasting frequencies included ARID1A and PCDH1 which were more common in gastric tumors and APC, BRAF, KRAS which were more common in colorectal tumors. These genes were identified as driver genes in gastrointestinal and other cancers (14,15). In summary, genetic mutations of gastrointestinal malignancies were heterogeneous across tumors and anatomical locations.
We identified two mutated genes, the E-cadherin CDH1 and the cell adhesion RHOA, accounting for the unique phenotypic features of gastric tumors: these mutated genes were highly specific to diffuse histology and advanced stages of gastric tumors. More importantly, the underlying genetic features of these mutations revealed that CDH1 and RHOA manifested differently in diffuse tumors as compared to intestinal tumors, and differently in gastric tumors as compared to colorectal and esophageal tumors. In diffuse-gastric tumors, we found one CDH1 recurrent hotspot involving G>T missense substitutions at nucleotide position 760 on exon number 6 which were known to impair the calcium binding pocket connecting the extracellular cadherin domains EC1 and EC2 leading to hereditary blepharocheilodontic syndrome (16). In addition, a large number of CDH1 mutations in diffuse-gastric tumors clustered in the sequence segment between nucleotide position 500 and 1,000 on exons 5, 6, and 7 which affect the extracellular cadherin domains EC1 and EC2 and the calcium binding pocket connecting these two domains. In contrast, there were no recurrent hotspots or clustered segments of CDH1 mutations in colorectal or esophageal tumors. The CDH1 gene codes calcium-dependent cell adhesion proteins which are involved in mechanisms regulating cell-cell adhesions, mobility, and proliferation of epithelial cells and has a potent invasive suppressor role (17). While the germline CDH1 has been known to account for hereditary diffuse-gastric cancer (18-20), this study shows that somatically mutated CDH1 also defines diffuse-gastric cancer.
We found two RHOA recurrent hotspots involving A>G missense substitutions at nucleotide position 125 on exon number 2 and T>G missense substitutions at nucleotide position 169 on exon number 3 in diffuse-gastric tumors. These hotspots were known to impair effector binding and GDP/GTP binding (21-23). Diffuse morphological phenotype is characterized by early breaking off of signet ring cells through the basement membrane, which requires resistance to anoikis, followed by the acquisition of highly infiltrative behavior; literature indicates that the ability of RHOA hotspot mutants to promote anoikis evasion in the organoid culture system is consistent with the critical role of RHOA in this process (21,22). It has long been known that diffuse-gastric cancer is often associated with advanced tumor stages (2,24-26). If the role of RHOA in fostering tumor cell survival is further confirmed, targeting the RHOA pathway may become useful in the treatment of diffuse-gastric cancer.
This study, to our knowledge, is one of the largest genetic analyses of gastrointestinal malignancies, making use of thirteen open-access cancer genomics datasets including nearly 2,000 subjects. Altogether, the genetic landscapes of CDH1 and RHOA mutations justified why the presence of these mutations placed diffuse-gastric cancer subjects at higher risk for advanced tumor spread than intestinal-gastric, colorectal, and esophageal cancer subjects. Our next step is to use this information to design therapeutic strategies to target CDH1 and RHOA mutant gastric tumors.
Acknowledgments
Funding: This work was partially supported by grant UL1 TR001414 from the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), through the Biostatistics, Epidemiology and Research Design Unit. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethics approval for this study was exempted by the Institutional Review Board at the University of California in Irvine because this study is a non-human subject research, making use of public datasets with non-identifiable subjects.
References
- Surveillance, Epidemiology, and End Results (SEER) Program () SEER*Stat Database: Cancer Stat Facts, Colorectal and Stomach Cancer, National Cancer Institute, DCCPS, Surveillance Research Program, released December 2018. Underlying mortality data provided by NCHS ().www.seer.cancer.gov
- Hoang T, Park M, Hiyama D, et al. Predictors of outcomes in patients with gastric cancer treated with contemporary multimodality strategies—a single institution experience. J Gastrointest Oncol 2019. [Crossref]
- Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand 1965;64:31-49. [Crossref] [PubMed]
- Hamilton SR, Aaltonen LA. editors. Tumours of the Digestive System. In: World Health Organization Classification of Tumours: Pathology and Genetics. Lyon, France: IARC Press, 2000.
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Tan IB, Ivanova T, Lim KH, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology 2011;141:476-85, 485.e1-11.
- Institutional Review Board at the University of California in Irvine. Available online: https://www.research.uci.edu/compliance/human-research-protections/researchers/activities-irb-review.html
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Grossman RL, Heath AP, Ferretti V, et al. Toward a shared vision for cancer genomic data. N Engl J Med 2016;375:1109-12. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995;57:289-300. [Crossref]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248-9. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Bailey MH, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;174:1034-5. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495-501. [Crossref] [PubMed]
- Ghoumid J, Stichelbout M, Jourdain AS, et al. Blepharocheilodontic syndrome is a CDH1 pathway-related disorder due to mutations in CDH1 and CTNND1. Genet Med 2017;19:1013-21. [Crossref] [PubMed]
- UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 2019;47:D506-15. [Crossref] [PubMed]
- Guilford P, Hopkins J, Harraway J, et al. E-cadherin germline mutations in familial gastric cancer. Nature 1998;392:402-5. [Crossref] [PubMed]
- Yoon KA, Ku JL, Yang HK, et al. Germline mutations of E-cadherin gene in Korean familial gastric cancer patients. J Hum Genet 1999;44:177-80. [Crossref] [PubMed]
- Yabuta T, Shinmura K, Tani M, et al. E-cadherin gene variants in gastric cancer families whose probands are diagnosed with diffuse gastric cancer. Int J Cancer 2002;101:434-41. [Crossref] [PubMed]
- Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573-82. [Crossref] [PubMed]
- Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014;46:583-7. [Crossref] [PubMed]
- Atlas of Genetics and Cytogenetics in Oncology and Haematology. Available online: http://AtlasGeneticsOncology.org
- Riquelme I, Saavedra K, Espinoza JA, et al. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget 2015;6:24750-79. [Crossref] [PubMed]
- Pernot S, Voron T, Perkins G, et al. Signet-ring cell carcinoma of the stomach: Impact on prognosis and specific therapeutic challenge. World J Gastroenterol 2015;21:11428-38. [Crossref] [PubMed]
- Kunz PL, Gubens M, Fisher GA, et al. Long-term survivors of gastric cancer: a California population-based study. J Clin Oncol 2012;30:3507-15. [Crossref] [PubMed]