
Discovery of a novel three-long non-coding RNA signature for predicting the prognosis of patients with gastric cancer
Introduction
Gastric cancer (GC), the fifth most common malignant tumor and the third leading cause of tumor related death, remains the public health burden globally. In 2018, there were more than 1,033,000 diagnosed cases and 782,000 GC-related mortalities worldwide (1). Despite diagnostic and therapeutic advances, the overall 5-year survival rate remains dismal. Therefore, it is urgently recommended to identify novel powerful biomakers of prognosis prediction and treatment response in patients with GC.
Long non-coding RNAs (lncRNAs) were regarded as RNA transcripts longer than 200 nt without the protein coding capability. Nowadays, lncRNAs have been considered to modulate gene expression at chromatin organizational, transcriptional and post-transcriptional levels (2,3), and thus take part in diverse tumorigenic processes including proliferation, anti-apoptosis and distant metastasis (4). Moreover, lncRNAs function as potential prognostic biomarkers and thus lncRNA-based signatures have been demonstrated in a wide range of malignancies via mining The Cancer Genome Atlas (TCGA). or GEO dataset (5-8). Several lncRNA-based predictive signatures for GC patients without the cross validation or validation in other cohorts have also been reported (9-11). Nevertheless, it is still necessary to establish a lncRNA-based model with high robustness and reliability in predicting prognosis of GC patients
We aimed to construct a predictive model of GC based on lncRNAs. RNA-Seq data of GC were obtained from TCGA to identify differentially expressed lncRNAs (DElncRNAs). Then, Robust likelihood-based survival model was employed to screen out candidate DElncRNAs with high robustness, followed by the multivariate Cox regression analysis in a stepwise manner. We present the following article in accordance with the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-140.
Methods
Data source and preprocessing
The level 3 seq-data of transcriptome as well as related clinical information of GC were downloaded from TCGA portal (up to Mar 26th, 2019), containing 375 GC and 32 adjacent paratumor tissues. To avoid confounding factors, only samples with follow-up time more than one month were incorporated in our study. Then, the remaining patients were randomly partitioned into the training and the testing set using the R “caret” package (Table 1). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Since all data were extracted from publicly available database, an approval by the ethics committee and patient consent were not required. We complied with the TCGA publication guidelines and data access policies.

Full table
Preliminary screening of differentially expressed lncRNAs
We employed “edger” R package to screen out DElncRNAs between GC and paratumor samples. In addition to the threshold of log2|fold change| >1 and FDR <0.05, lncRNAs were also filtered as the following criteria: first, mean expression of each lncRNA in all samples >1; second, expression of each lncRNA in more than 50% samples >1. Third, the variance of expression level of each lncRNA was higher than 20% of the total expression variances of all lncRNAs.
Discovery and selection of prognosis related lncRNAs
The relationship between DElncRNAs and overall survival (OS) was first analyzed in the training set. The univariate Cox regression analysis of the screened DElncRNAs was performed using R “survival” package. Those with P value <0.05 were regarded as candidate lncRNAs.
To construct a lncRNA-related prognostic signature, the Robust likelihood-based survival model was performed using R “rbsurv” package as follows:
- We randomly partitioned patients into the training set with N×(1 − p) and the validation set with N×p, p = 1/3, fitted each lncRNA to the training set and obtained a parameter value, the log-likelihood of which was further confirmed in the validation set.
- We repeated the above procedure 10 times and obtained 10 log-likelihoods for each lncRNA. The one with the highest log likelihood was considered as the best lncRNA. We searched the next best lncRNA by evaluating each two-lncRNA model and obtained the optimal one with highest mean log-likelihood.
- A large number of models emerged through the above procedure. We calculated Akaike information criterions (AICs) for all the candidate signatures and constructed the optimal one with the smallest AIC.
Construction and validation of the lncRNA-based predictive signature
Candidate LncRNAs among the optimal model were then incorporated into the stepwise Cox multivariate analysis in the training set. A prognostic signarue was finally established by the multiplication of lncRNA expression (X) and regression coefficient (β). Then, each patient was scored in accordance with the following formula: Risk Score=X1β1+X2β2+.+Xnβn.
To confirm the predictive performance of the lncRNA-based model, a receiver operating characteristic (ROC) was constructed using the R “survivalROC” package, from which a best cutoff value with the maximal sensitivity and specificity was obtained. Using the optimal cutoff derived from the training set, patients from three cohorts were divided into the high- and low-risk group, respectively. Kaplan-Meier survival curve with log-rank test analysis was employed to compare OS between high- and low-risk groups.
LncRNA-based signature for prediction independent of other clinical features
The lncRNA-based model together with other clinical features including age, gender, TNM stage were referred to the univariate Cox regression model. Then, factors associated with OS were incorporated into the multivariate Cox regression model to determine whether the lncRNA-based signature was an independent prognostic indicator of OS.
Enrichment analysis of the three lncRNA-based signature
Gene set enrichment analysis (GSEA) utilizes predefined gene sets to sort related genes in accordance with the extent of difference between the two types of samples, and then to evaluate whether gene sets are at the top or bottom of the sorting table. Herein, we utilized GSEA to screen out potential pathways between high- and low-risk group stratified by the best cutoff from the training set. Molecular signatures included in our study contains C2 curated gene set and C5 GO gene set. Enrichment score (ES) was obtained using permutation analysis 1000 times. P<0.01 was considered statistically different.
Results
Identification of prognosis related lncRNA
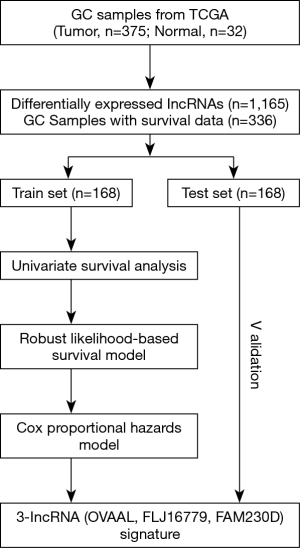
We performed our study as described in the flow chart (Figure 1). Using the R “edgeR” package, we identified a total of 1,165 DElncRNAs (|log2FC| >1, FDR <0.05) between 375 GC and corresponding 32 non-tumor samples, including 904 up-regulated and 261 down-regulated DElncRNAs, which were subjected to the univariate Cox regression model in the training set, and finally only 202 prognoses related DElncRNAs were selected for further study. Moreover, the unsupervised hierarchical cluster analysis of the top 20 DElncRNAs sorted by FC was performed and visualized by R “pheatmap” package (Figure S1).
Establishement of risk score system based on the 3-lncRNA signature
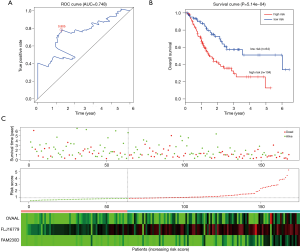
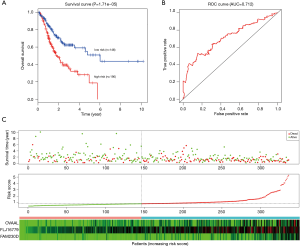
Robust likelihood-based survival analysis of 202 prognosis-related DElncRNAs were conducted using the R “rbsurv” package, followed by the Cox multivariate regression model. Thus, we identified a series of candidate lncRNAs with high frequency which were further incorporated into the stepwise Cox multivariate model, and a prognostic signature consisting of three lncRNAs (OVAAL, FLJ16779, FAM230D) was finally constructed, all of which were considered as risky factors. The risk score of each GC sample was computed in accordance with the 3-lncRNA risk formula. Additionally, the ROC curve of the training set indicated that the optimal cutoff was 0.805 (Figure 2B).
Predictive performance of the 3-lncRNA model for GC
In the training set, we classified GC patients into high-risk and low-risk group using the optimal cutoff of risk scoring system. As can be seen from Figure 2A, patients in the high-risk group suffered remarkably poorer OS than those from the low-risk group, the 5-year OS was 12.8% and 51.0%, respectively. The area under curve (AUC) of time-dependent ROC, indicative of reliability, was 0.748 at 5-year (Figure 2B). When GC patients were sorted in ascending order of the risk score, the expression pattern of all three lncRNAs increased accordingly, and the mortality of patients in the high-risk group was much higher than those in the low-risk group (Figure 2C).
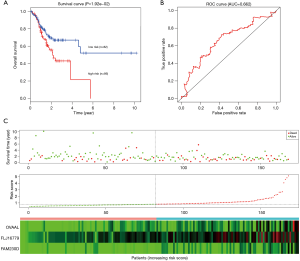
To determine the robustness of survival prediction in GC, 3-lncRNA model was validated in the test and entire set. Using the optimal cut off derived from the training set, GC patients in other two cohorts were also divided into high- and low-risk groups. In the testing set, patients in the high-risk group had notably OS than those in the low-risk group. The 5-year OS was 21.5% and 52.0% (Figure 3A), respectively. The 5-year AUC of time-dependent ROC was 0.622 (Figure 3B). The distributions of survival status, risk score and expression profiles among patients were described in the Figure 3C. Similar results of survival prediction and reliability were also observed in the entire set. The 5-year OS in the high- and low-risk group was 18.7% and 50.9% (Figure 4A), respectively. The 5-year AUC of time-dependent ROC was 0.712 (Figure 4B). When patients were sorted in ascending order of the risk score, we found the same trends of survival status and expression pattern in the testing (Figure 3C) and entire set (Figure 4C) as described in the training set.
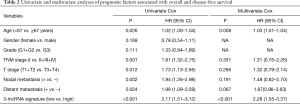
The predictive power of the 3-lncRNA model independent of other clinical variables
The univariate Cox regression model indicated that the 3-lncRNA model was a poor prognostic factor for OS. To explore its predictive role independent of clinical variables, the 3-lncRNA model together with other clinical features were incorporated into the multivariate Cox regression analysis. As a result, the three-lncRNA model served as an independent poor prognostic indicator for OS (Table 2).
Full table
Identification of the 3-lncRNA related functions and pathways
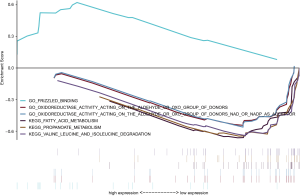
In order to explore the potential mechanism of lncRNA promoting the occurrence and development of gastric cancer, we performed GSEA in high- and low-risk group of all GC patients using KEGG (C2) and Molecular function of GO (C5) signatures. In the high-risk group, eight GO terms were significantly clustered, but no KEGG pathways were identified (P<0.05). In the low risk group, ten KEGG pathways, mainly focusing on metabolism and apoptosis, and 22 GO terms were significantly enriched (P<0.05). Gene sets with P<0.01 were selected and presented as a multi-GSEA enrichment plot (Figure 5).
Discussion
GC remains a major public health burden worldwide especially in northeast Asian countries including China. Compared to colorectal cancers, GC patients underwent a poorer prognosis due to the high risk of relapse and metastasis. Thus, it is urgently recommended to identify a novel prognostic biomarker of GC. And the Combined gene signature may better the predictive efficacy than the single biomarker. Recently, gene signatures composed of several lncRNAs have gained more attention and play a pivotal role in prognosis prediction of malignancies (5-8).
Here, we constructed a novel 3-lncRNA (OVAAL, FLJ16779, FAM230D) prognosis predictive model for GC in the training set and then validated its accuracy and robustness in other two cohorts. Moreover, the 3-lncRNA model was an independent prognostic predictor for GC and patients in the high-risk group underwent poorer OS than those in the low-risk group. In general, the risk model composed of the 3 lncRNAs could be an effective predictor for GC prognosis and even improve the prognosis. The patients with high risk scores should be followed up more frequently for relapse and metastasis. In addition, GSEA revealed several gene sets from diverse molecular signatures respectively enriched in the high- or low-risk group, which might account for the underlying mechanism of the 3-lncRNA based signature. The Frizzled binding, one of enriched Gos, may induces the initiation and progression of GC via transmiting extracellular signals. Frizzled receptors belong to G protein coupled receptors (GPCRs) which function in Wnt signaling. Thus, Frizzled receptors are essential for embryonic development, maintenance of stem cell self-renewal, and its aberrant expression correlates with multiple cancers (12,13).
Three lncRNAs indentified in our study have not been documented to be biomarkers with prognostic role in other malignancies. Ovarian Adenocarcinoma Amplified LncRNA (OVAAL), also known as OVAL or LINC01131 (14), was first reported to confer apoptotic resistance in multiple cancer types (15). Compared to corresponding adjacent tissues, OVAAL expression was markedly up-regulated in colorectal cancer and melanoma. Functional and mechanistic investigations indicated that knockdown of OVAAL increased proliferation and inhibited apoptosis through physically interacting with serine/threonine-protein kinase 3 (STK3) and then activating the downstream RAF/MEK/ERK pathway. Similarly, OVAAL was also identified as an aberrantly high expressed and a prognosis related lncRNA in our study. Moreover, GC patients were classified into high- and low-expression group in accordance with the median value of OVAAL and then were subjected to the GSEA. Elevation of OVAAL was associated with Dorsal-ventral axis formation and melanoma (Figure S2A), which might provide novel molecular mechanism of OVAAL in the pathogenesis and progression of GC.
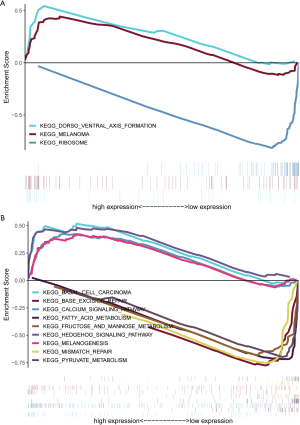
Little information is available for FLJ16779 and FAM230D, which prompted us to evaluate the function and mechanism of these lncRNAs using GSEA. Surprisingly, a total of 11 and 54 gene sets were enriched in the high- and low-expression of FLJ16779 group (P<0.01) (data not shown). Activated gene sets were mainly clustered in tumor related pathways or carcinogenesis, such as Hedgehog signaling pathway, Calcium signaling pathway, Basal cell carcinoma and Melanogenesis. While repressed gene sets were primarily associated with metabolism including Carbohydrate, fat acid and amino acid, as well as several gene repair pathways (Figure S2B). In fact, reprogramming of energy metabolism has already been recognized as a hallmark of cancer (16), and lncRNAs play a crucial role in the biological process (17). These results showed that the prognosis related FLJ16779 was implicated in gastric carcinogenesis and progression via modulating energy metabolism. In addition, FAM230D belongs to a family of long intergenic non-coding RNA genes, which locate in human chromosomal region 22q11.2 and carry a DNA translocation breakpoint/AT-rich sequence (18). Unfortunately, we failed to predict the potential role of FAM230D in gastric tumorigenesis whether utilizing GSEA or the method of co-expressing genes. Perhaps, FAM230D facilitates the progression of GC dependent on the other two functional lncRNAs.
Nevertheless, some limitations may exist in our study. First, there were several invalid data regarding the follow-up information in TCGA. Although patients with the follow-up time less than 1 month were removed, the 5-year OS of GC was 36.0%, slightly higher than 31.5% reported by the American Cancer Society (19). Second, the prognostic value of the three lncRNA signature should be verified in other internal cohorts despite cross-validation in our study.
Conclusions
In general, we established a novel three-lncRNA model by mining the TCGA dataset. The predictive module could not only function as a novel biomarker for the risk stratification of GC patients, but also shed light on how GC develops.
Acknowledgments
Funding: This work was supported by National Natural Scientific Foundation of China (No.81660415) and Natural Scientific Foundation of Inner Mongolia (No.2016MS0835).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-140
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-140). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Since all data were extracted from publicly available database, an approval by the ethics committee and patient consent were not required. We complied with the TCGA publication guidelines and data access policies.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Mao X, Qin X, Li L, et al. A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol Oncol 2018;149:181-7. [Crossref] [PubMed]
- Fan Q, Liu B. Discovery of a novel six-long non-coding RNA signature predicting survival of colorectal cancer patients. J Cell Biochem 2018;119:3574-85. [Crossref] [PubMed]
- Yan J, Zhou C, Guo K, et al. A novel seven-lncRNA signature for prognosis prediction in hepatocellular carcinoma. J Cell Biochem 2019;120:213-23. [Crossref] [PubMed]
- Zhang H, Zhu M, Du Y, et al. A Panel of 12-lncRNA Signature Predicts Survival of Pancreatic Adenocarcinoma. J Cancer 2019;10:1550-9. [Crossref] [PubMed]
- Cheng P. A prognostic 3-long noncoding RNA signature for patients with gastric cancer. J Cell Biochem 2018;119:9261-9. [Crossref] [PubMed]
- Miao Y, Sui J, Xu SY, et al. Comprehensive analysis of a novel four-lncRNA signature as a prognostic biomarker for human gastric cancer. Oncotarget 2017;8:75007-24. [Crossref] [PubMed]
- Ren W, Zhang J, Li W, et al. A Tumor-specific prognostic long non-coding RNA signature in gastric cancer. Med Sci Monit 2016;22:3647-57. [Crossref] [PubMed]
- Nusse R, Clevers H. Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 2017;169:985-99. [Crossref] [PubMed]
- Yang S, Wu Y, Xu TH, et al. Crystal structure of the Frizzled 4 receptor in a ligand-free state. Nature 2018;560:666-70. [Crossref] [PubMed]
- Akrami R, Jacobsen A, Hoell J, et al. Comprehensive analysis of long non-coding RNAs in ovarian cancer reveals global patterns and targeted DNA amplification. PLoS One 2013;8:e80306. [Crossref] [PubMed]
- Sang B, Zhang YY, Guo ST, et al. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc Natl Acad Sci U S A 2018;115:E11661-70. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Shankaraiah RC, Veronese A, Sabbioni S, et al. Non-coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett 2018;419:167-74. [Crossref] [PubMed]
- Delihas N. A family of long intergenic non-coding RNA genes in human chromosomal region 22q11.2 carry a DNA translocation breakpoint/AT-rich sequence. PLoS One 2018;13:e0195702. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]


