
An alternative splicing signature model for predicting hepatocellular carcinoma-specific survival
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the world (1). The clinical effect of individualized treatment is still unsatisfactory, as individual therapy strategies and optimal management still pose significant challenges. Surgical resection is presently the only possible treatment for HCC (2). Unfortunately, as newly diagnosed HCC patients present with distant metastases, only a few patients can undergo radical surgery. More seriously, many HCC patients will develop recurrence, mostly within 2 years (3). Evidence accumulated during the past few years indicates that tumor emergence, recurrence, and metastasis of HCC is a complex and rigorous regulatory process (4). These changes not only activate oncogenes or inhibit the role of tumor suppressor genes, but also endow HCC with the ability to invade and metastasize (5). In this way, further understanding the relationship between the biological mechanism of HCC and the corresponding clinicopathological characteristics is an important step towards conducting targeted therapy and improving the prognosis of patients with HCC.
The rapid development of the high-throughput technology marks a new era of cancer genomics study (6). In recent years, the application of RNA sequencing and microarray technology (7), along with the genomic profiling of HCC, has been sufficiently researched, while studies examining RNA expression, copy number variation (CNV), and DNA methylation have been widely performed. More importantly, these results have further confirmed that multiple genetic events are required to promote the malignant progression of HCC (8,9), and that the complexity of cancer biology can only be understood under a genome-wide schema. However, although these studies have provided valid findings, they have mainly focused on the transcriptional expression level (10,11), while the systematic analysis of transcript architecture variation has been greatly ignored, especially in alternative splicing (AS).
AS refers to a process in which an RNA precursor can be spliced into different segments to produce different messenger RNA (mRNA) with different structure and function, which is then used to form protein variants. It is one of the most widely applied mechanisms that can account for proteomic diversity and cellular complexity (12), and is a major mechanism of protein diversity. There is growing evidence that splicing dysregulation is associated with cancer (13). From this point of view, aberrant AS events can directly affect the occurrence and progression of cancer. Moreover, a growing amount of evidence suggests that unbalanced or incorrect expression of AS is another biomarker of cancer (14,15). Thus, cancer-related AS events can potentially be used as therapeutic targets and diagnostic and predictive biomarkers. We assume that AS is also involved in the pathogenesis of HCC, and may thus be a potential prognostic factor.
The rapid accumulation of RNA sequence data in clinical samples makes it possible to study the role of AS in clinical outcomes in relatively large populations. Here, we systematically analyzed the genome-wide AS events of HCC patients in The Cancer Genome Atlas (TCGA). Our results revealed that HCC survival-related AS events are important in HCC, can affect the progression of HCC, and can be used as highly efficient and reliable prognostic signatures for HCC patients. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-377).
Methods
Data collection
Clinical information of HCC was downloaded from TCGA web site (https://portal.gdc.cancer.gov/) (16), and the publication guidelines provided by TCGA were followed. AS events of HCC were downloaded from TCGA SpliceSeq web site (17). The website presents gene splicing patterns and related statistical data in an intuitive, interactive, and graphical form, and has dynamic characteristics for exploring splicing variations across tumor types. The data query or download function of TCGA SpliceSeq’s data enabled us to retrieve AS data for each sample and stitching event, which were then included in our comprehensive analysis.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data processing
Some patients were excluded from analysis for the following reasons: (I) no histological diagnosis of HCC, (II) incomplete clinical survival data, or (III) no corresponding AS event data. For each transcript, the relative abundance of each sample, which we called “proportion spliced-in” (PSI) (18), was calculated by standardizing the sum of its abundance in each transcript per million units in the same gene. PSI value has been habitually used to quantify AS events (19). PSI was rated from 0% to 100%. To generate a reliable series of AS events, we implemented strict filters: (I) an average PSI value ≥0.05, (II) a percentage of samples with PSI value ≥75, (III) a mean standard deviation (SD) of PSI value ≥0.001. The HCC patients were separated into two groups by the median cut. To describe a splicing event precisely, every splicing event was presented by a unique code in the current study. For example, for the code, UBB-39434-RI, UBB is the gene name, 39434 is the order number of the splicing event in the dataset, and RI is the type of AS.
Statistical analysis and software
The Kaplan-Meier survival analysis was used to analyze the survival time of HCC patients. The AS events with a P value <0.05 from Kaplan-Meier analysis were identified as survival-related AS events. Circle maps were created to show the details of these AS events, and included P value, hazard ratio (HR), and 95% confidence interval (CI). Multivariate Cox proportional hazards regression analysis was used to select the survival-related AS events that might be independent predictors for HCC patient prognosis. Receiver operating characteristic (ROC) curves were used to show the predictive efficiency of the prognostic models. All statistical analyses in this study were performed using R software (version 3.5.2) and SPSS Statistics, version 24.0 (SPSS Inc., Chicago, IL, USA). A two-tailed, P value <0.05 was considered to indicate a statistically significant difference.
Results
AS events in HCC
SpliceSeq is a resource for RNA sequence data from TCGA. It provides an overview of AS events and identifies splicing events with potential functional changes caused by splice variations. We generated AS profiles in HCC patients in TCGA. In this cohort, we identified 78,878 AS events from 13,045 genes. On the basis of the splicing pattern, AS events can be divided into seven types (20): including alternate acceptor site (AA), alternate donor site (AD), alternate promoter (AP), alternate terminator (AT), exon skip (ES), mutually exclusive exons (ME), and retained intron (RI) (Figure 1A).
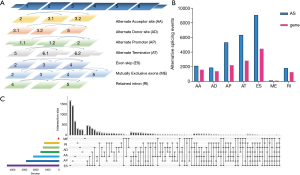
Most AS events can only be detected in a few samples (21). In addition, the expression level of some splicing isoforms is very low and excludes individual influence. In order to generate AS event data as reliably as possible, we implemented a series of strict filters. We obtained 26,495 as events from 8,182 genes after screening, which indicated that one gene may have one or more AS events (Figure 1B).
We use Venn diagrams to represent the relationships between interaction sets traditionally, but when dealing with five or more sets, it can be extremely complicated. Thus, we used the UpSet plot (22), a creative visualization technology to quantitatively analyze the interaction sets, to analyze the intersection of seven types of AS events. As shown in Figure 1C, most of the AS events and genes did not have a one-to-one correspondence, and one gene might have up to seven types of AS events. For example, the ACSL1 gene had the AP, ES, and ME types of splicing, while the PTK2 gene had all the seven types of splicing. Different combinations of genes and AS events may provide the greatest potential process to enrich the diversity of the transcriptome.
Identifying survival-related AS events in HCC
Clinical parameters of the HCC patients’ cohort were also downloaded from TCGA database. We collected 319 HCC patients with disease-free survival (DFS) data and 367 HCC patients with OS data in this study. These were named the DFS cohort and the OS cohort, respectively.
In the DFS cohort, the median DFS was 20.9 months. Furthermore, the 1-, 2-, and 5-year DFS rates were 64.3%, 45.8%, and 28.0%, respectively. For each AS event, the PSI value was divided into two groups according to the median cut. We detected a total of 2,440 survival-related AS events in the DFS cohort (http://fp.amegroups.cn/cms/9ced11df84d805614c16a456f3a2edc1/jgo-20-377-1.xlsx). The top 15 survival-related AS events in each type of AS are presented in Figure 2. The survival-related AS events had different prognostic values. For example, more than half of the top 15 survival-related AS events and RI events indicated a bad prognosis for HCC patients, while 13 of the top 15 ME events indicated a good prognosis for HCC patients.

In the OS cohort, the median OS was 55.7 months. The 1-, 2-, and 5-year OS rates were 82.5%, 69.6%, and 46.0%, respectively. For each AS event, the PSI value was also divided into two groups according to the median cut. We detected a total of 2,888 survival-related AS events in the OS cohort (http://fp.amegroups.cn/cms/e4f99f2a3e0feec57647eb416228df17/jgo-20-377-2.xlsx). The top 15 survival-related AS events in each type of AS are presented in Figure 3. The survival-related AS events had different prognostic values; in particular, 14 of the top 15 ES events indicated a good prognosis for HCC patients.

Building the prognostic model for HCC patients
The relationship between the AS events we identified, and the prognosis of HCC patients was studied in TCGA cohort. Univariate survival analyses of OS and DFS were then respectively conducted. We selected the top significant survival-related AS events in the seven types as the candidates. We excluded any event that might not have been an independent factor in the prognosis model, and binomial logistic regression was applied to the potential AS events in the seven AS types.
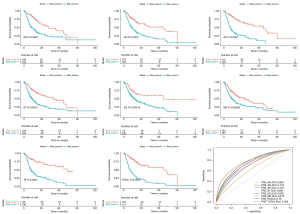
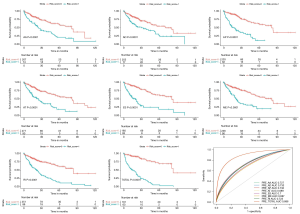
In both the DFS and OS cohort, the HCC patients were classified into the low-risk and the high-risk groups (Figures 4,5). We used the median value of the risk score as the cutoff point. In the prognostic model based on the seven different types of AS, the survival time of the two subgroups was significantly different (P<0.05). These showed great power in distinguishing bad or good outcomes in HCC patients. Then, the candidate predictors from seven AS types were combined and further analyzed to build the prognostic models for HCC patients. The equations are listed below. In the DFS model, the median DFS of the low-risk and the high-risk groups was 20.7 and 1.8 months, respectively. In the OS model, the median OS of the low-risk and the high-risk groups was 83.2 and 14.9 months, respectively.
The risk score model of DFS was calculated as follows: Logit P = (CES1 − 655795 − AA × − 0.913) + (TOP1MT − 85417 − AA × −0.978) + (C6orf1 – 75778 − AD × 1.241) + (TAF6 – 80899 − AD × 1.209) + (CYP2R1 − 14478-AP × −0.804) + (ATAD3B − 173 − AT × −0.872) + (MRPL52 – 26646 – RI × 1.267).
The risk score model of OS was calculated as follows: Logit P = (MAP7D1 − 1758 − AA × −0.582) + (RNF19B – 1647 − AA × −0.988) + (PLEKHH3 − 41103 − AD × −1.204) + (OSGIN1 − 37798 − AP × −0.766) + (FAM107B − 10823 − AP × − 1.031) + (OGFOD2 –25005 − AP × 0.94) + (GPR75 − ASB3 − 53555 − AP × − 0.782) + (ADRA1A − 83140 − AT × −0.713) + (BBS9 – 79224 − ES × −1.124) + (RAB6A − 17707 − ME × 0.857) + (UBB – 39434 − RI × 0.847).
The ROC curves were generated to compare the efficiencies of the two predictive models. We calculated and compared the area under the curve (AUC) of each predictive model. As can be seen in Figure 4 and Figure 5 shown, the prognostic model with a total of seven types of AS events had the highest efficiency in distinguishing bad or good outcomes of HCC patients. The AUC of the DFS cohort’s prognostic predictor with a total of seven types was 0.769, and the AUC of the OS cohort’s was 0.886. The detailed information of AS events in the prognostic model is listed in Table 1 and Table 2.
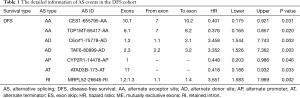
Full table
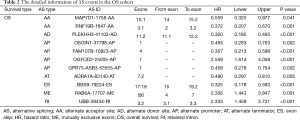
Full table
Discussion
AS is one of the post-transcriptional modifications in which exonic or intronic regions are removed from the precursor mRNA (23). Through the splicing of exons together to produce mature RNAs, it is critical source of protein species diversity (24). In other words, changes in AS events may lead to changes in protein functions and the life activities based on these functions (25). In recent years, increasing evidence has shown that AS is closely related to the occurrence of cancer (26). The cancer cells often take advantage of the process of AS to promote cancer cell proliferation, metastasis, and even drug resistance. More importantly, a growing amount of data from genome-wide studies indicate that more than 90% of human genes have experienced AS events (27-29). These outcomes demonstrate that not only gene expression, but also AS subtypes need to be better investigated.
HCC is induced by many pathogenic factors, with infection by hepatitis, alcoholism, or aflatoxin B1 exposure being particularly common (30). Hepatocarcinogenesis is a complicated and multi-step process, which includes the unbalanced expression of AS variants or the incorrect expression of the subtypes. A growing body of research suggests that AS plays an important regulatory role in HCC; splicing subtypes such as TP53, K-ras, and L-myc are all expressed, and the functional characteristics of AS events are obvious and include antitumor effects. Furthermore, the expression of cell-fate determinant Numb was found to be abnormal in HCC, and Numb is alternatively spliced. One AS subtype of Numb contains a long proline-rich region (PRRL) and the other contains a short proline-rich region (PRRS). According to reports, Numb PRRL is associated with early postoperative recurrence and reduced overall survival (31).
Deeper research into AS might provide specific insights into the mechanisms of HCC. It is reported that overexpression of SRSF2 is involved in the occurrence and development of HCC. Further studies have shown that cancer-related splicing variants upregulated by SRSF2 in HCC clinical specimens are essential for the pathogenesis and progression in HCC cells (32). Similarly, The CD44 standard isoform is involved in epithelial–mesenchymal transition in HCC, and the expression levels of CD44 variant-containing exons, v5, v6, v7–8, and v10, are correlated with a high histological grade of HCC (33). Moreover, the elevated expression of CD44 v6 is associated with the vascular invasion of HCC tissues and invasive potentials in HCC cell lines.
Previous studies on AS in HCC have mainly focused on identifying cancer-specific AS events by comparing cancer tissues or cells with healthy counterparts (34,35). However, the survival-related AS events in HCC have remained largely unstudied. In this study, we systematically identified and analyzed the survival-related AS events in HCC patients from TCGA database. A total of 78,878 AS events from 13,045 genes were detected. There were 2,440 and 2,888 AS events found to be significantly associated with the DFS and OS of HCC patients. The prognostic genes included CYP2R1, ATAD3B, OSGIN1, FAM107B, and ADRA1A, which play vital roles in HCC. Among the models constructed based on all seven types, which included the AA-type, AD-type, AP-type, AT-type, ES-type, ME-type, and RI-type models, the final risk score model showed an impressive efficiency in predicting the prognosis of HCC patients. The risk score model built in our study also performed well in HCC. The AUCs of ROC were 0.769 for DFS and 0.886 for OS. The results above indicate that AS not only has important biological functions but also has potential clinical value in HCC.
Some limitations to our study should also be addressed. For one, much of TCGA SpliceSeq data only contains AS events of protein-coding genes and do not include noncoding RNAs, which also have splicing patterns that play an important role in cancer progression (34). Furthermore, molecular analysis of HCC is often challenging because each tumor sample can be polyclonal, and different samples contain different levels of stromal tissue (36). This study is a computational work based on TCGA SpliceSeq data (37). Despite its limitations, the TCGA SpliceSeq database provides a standardized data set from which we can derive main clinical outcome endpoints and solutions to issues. As discussed above, we systematically profiled the genome-wide AS events in HCC samples from TCGA, identified the survival-related AS events, and investigated their prognostic value. Our results reveal that such survival-related AS events are important in HCC, can directly affect the progression of HCC, and can be used as a reliable and efficient prognostic signature for HCC patients. We consider these findings to be reliable and valuable for HCC prognosis.
In conclusion, we found the survival-related AS events to be an ideal prognostic predictor, and our final model performed well in risk stratification in HCC patients. Among these AS events, several valuable therapeutic targets were identified for future verification and may provide further insights into the underlying mechanism of AS in hepatocarcinogenesis.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-377
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-377). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Morise Z. Laparoscopic liver resection for the patients with hepatocellular carcinoma and chronic liver disease. Transl Gastroenterol Hepatol 2018;3:41. [Crossref] [PubMed]
- Du ZG, Wei YG, Chen KF, et al. Risk factors associated with early and late recurrence after curative resection of hepatocellular carcinoma: a single institution's experience with 398 consecutive patients. Hepatobiliary Pancreat Dis Int 2014;13:153-61. [Crossref] [PubMed]
- Akateh C, Pawlik TM, Cloyd JM. Adjuvant antiviral therapy for the prevention of hepatocellular carcinoma recurrence after liver resection: indicated for all patients with chronic hepatitis B? Ann Transl Med 2018;6:397. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref] [PubMed]
- Idris SF, Ahmad SS, Scott MA, et al. The role of high-throughput technologies in clinical cancer genomics. Expert Rev Mol Diagn 2013;13:167-81. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell 2014;5:673-91. [PubMed]
- Ning L, Wentworth L, Chen H, et al. Down-regulation of Notch1 signaling inhibits tumor growth in human hepatocellular carcinoma. Am J Transl Res 2009;1:358-66. [PubMed]
- Zang HL, Ren SN, Cao H, et al. The ubiquitin ligase TRIM25 inhibits hepatocellular carcinoma progression by targeting metastasis associated 1 protein. IUBMB Life 2017;69:795-801. [Crossref] [PubMed]
- He JH, Han ZP, Liu JM, et al. Overexpression of Long Non-Coding RNA MEG3 Inhibits Proliferation of Hepatocellular Carcinoma Huh7 Cells via Negative Modulation of miRNA-664. J Cell Biochem 2017;118:3713-21. [Crossref] [PubMed]
- Blencowe BJ. Alternative Splicing: New Insights from Global Analyses. Cell 2006;126:37-47. [Crossref] [PubMed]
- Feng H, Qin Z, Zhang X. Opportunities and Methods for Studying Alternative Splicing in Cancer with RNA-Seq. Cancer Lett 2013;340:179-91. [Crossref] [PubMed]
- Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int J Cell Biol 2013;2013:463786. [Crossref] [PubMed]
- Kitamura K, Matsushita K, Kobayashi S, et al. Alternative Splicing Detection as a Biomarker for Cancer Diagnosis: A Novel Progressive Mechanism of Acute Lymphoblastic Leukemia with Alternative Splicing as a Biomarker Candidate. Rinsho Byori 2015;63:1091-102. [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-16.e11. [Crossref] [PubMed]
- Ryan MC, Cleland J, Kim R, et al. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 2012;28:2385-7. [Crossref] [PubMed]
- Pervouchine DD, Knowles DG, Guigó R. Intron-centric estimation of alternative splicing from RNA-seq data. Bioinformatics 2013;29:273-4. [Crossref] [PubMed]
- Schafer S, Miao K, Benson CC, et al. Alternative Splicing Signatures in RNA-seq Data: Percent Spliced in (PSI). Curr Protoc Hum Genet 2015;87:11.16.1-11.16.14.
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet 2002;30:13-9. [Crossref] [PubMed]
- Romero JP, Muniategui A, De Miguel FJ, et al. EventPointer: an effective identification of alternative splicing events using junction arrays. BMC Genomics 2016;17:467. [Crossref] [PubMed]
- Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017;33:2938-40. [Crossref] [PubMed]
- Lopez AJ. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu Rev Genet 1998;32:279-305. [Crossref] [PubMed]
- Graveley BR. Alternative Splicing: Increasing Diversity in the Proteomic World. Trends Genet 2001;17:100-7. [Crossref] [PubMed]
- Climente-González H, Porta-Pardo E, Godzik A, et al. The Functional Impact of Alternative Splicing in Cancer. Cell Rep 2017;20:2215-26. [Crossref] [PubMed]
- Xiong Y, Deng Y, Wang K, et al. Profiles of alternative splicing in colorectal cancer and their clinical significance: A study based on large-scale sequencing data. EBioMedicine 2018;36:183-95. [Crossref] [PubMed]
- Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008;40:1413-5. [Crossref] [PubMed]
- Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008;456:470-6. [Crossref] [PubMed]
- Lin JC. Impacts of Alternative Splicing Events on the Differentiation of Adipocytes. Int J Mol Sci 2015;16:22169-89. [Crossref] [PubMed]
- Serio I, Napoli L, Leoni S, et al. Direct antiviral agents for HCV infection and hepatocellular carcinoma: facts and FADs. Transl Cancer Res 2019;8:S223-32. [Crossref]
- Lu Y, Xu W, Ji J, et al. Alternative splicing of the cell fate determinant Numb in hepatocellular carcinoma. Hepatology 2015;62:1122-31. [Crossref] [PubMed]
- Luo C, Cheng Y, Liu Y, et al. SRSF2 Regulates Alternative Splicing to Drive Hepatocellular Carcinoma Development. Cancer Res 2017;77:1168-78. [Crossref] [PubMed]
- Endo K, Terada T. Protein expression of CD44 (standard and variant isoforms) in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, p53 expression, and patient survival. J Hepatol 2000;32:78-84. [Crossref] [PubMed]
- Zhang L, Liu X, Zhang X, et al. Identification of important long non-coding RNAs and highly recurrent aberrant alternative splicing events in hepatocellular carcinoma through integrative analysis of multiple RNA-Seq datasets. Mol Genet Genomics 2016;291:1035-51. [Crossref] [PubMed]
- Wang XQ, Luk JM, Leung PP, et al. Alternative mRNA splicing of liver intestine-cadherin in hepatocellular carcinoma. Clin Cancer Res 2005;11:483-9. [PubMed]
- Friemel J, Rechsteiner M, Frick L, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res 2015;21:1951-61. [Crossref] [PubMed]
- Ryan M, Wong WC, Brown R, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res 2016;44:D1018-22. [Crossref] [PubMed]


