
Long non-coding RNA small nucleolar RNA host gene 7 facilitates the proliferation, migration, and invasion of esophageal cancer cells by regulating microRNA-625
Introduction
Esophageal cancer (EC) is an extremely aggressive malignancy with high rates of morbidity and mortality (1). There are 2 types of EC, esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC) (2). In China and other East Asian countries, ESCC accounts for as many as 90% of EC cases (3). Due to the lack of symptoms or signs in early ESCC, timely diagnosis is difficult, and many patients are already in the late stage of the disease when they first see a doctor. Despite advanced progress having been achieved in the clinical treatment of ESCC in recent years, the disease’s poor prognosis indicates that obstacles hindering successful recovery still exist (4). The pathogenesis of EC is extremely complex and involves many abnormalities in genes and signaling pathways. Therefore, exploration of the key signaling network of ESCC and its related molecular regulatory mechanisms is crucial for the discovery of effective therapeutic targets.
In recent years, the development of high-throughput sequencing technology has shifted researchers’ attention to long non-coding RNAs (lncRNAs). LncRNAs are RNA molecules with a transcript length longer than 200 nucleotides, the protein-coding potential of which are limited (5). More and more studies have found that lncRNAs, as multi-gene transcription and post-transcriptional regulatory factors, carry out important functions in the processes of liver cancer, lung cancer, lymphoma, gastric cancer, and many other malignancies (6). Many studies have evidenced the relationship between lncRNAs and the development of ESCC. For instance, lncRNA FAM83H-AS1 is significantly up-regulated in ESCC tissues. It has been found to be related to TNM staging, pathological differentiation, as well as lymph node metastasis, and to promote ESCC progression through the miR-10a-5p/Girdin axis (7). Meanwhile, the expression level of lncRNA CASC2 is obviously decreased in ESCC and is positively correlated with patients’ overall survival. More specifically, CASC2 can inhibit the proliferation, migration and invasion of ESCC cells via elevating SOCS1, and enhances drug sensitivity (8).
Small nucleolar RNA host gene 7 (SNHG7) is a newly identified lncRNA, which located on chromosome 9q34.3 and has a full length of 984 bp. It has been confirmed to be an oncogene in multiple tumors including glioblastoma, lung cancer, breast cancer, and hepatocellular carcinoma (9-12). In non-small cell lung cancer, SNHG7 can inhibit microRNA (miRNA/miR)-181a-5p via the Akt/mTOR signaling, and facilitate cell proliferation, migration, and invasion (9). SNHG7 can also promote the proliferation, migration and invasion of glioblastoma cells through its regulation of miR-449b-5p and its target gene MYCN (10). However, further study is needed to shed light on the expression, functions, and mechanism of SNHG7 in ESCC. Therefore, in the present research, we aimed to investigate the expression level of SNHG7 in ESCC tissues and cell lines, study the influences of SNHG7 on ESCC cell proliferation, migration, and invasion in vitro, and explore the corresponding molecular mechanisms. We present the study in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jgo-21-147).
Methods
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Nanyang Central Hospital Affiliated Hospital of Henan University (No. 20160075), and the specimens were obtained with the informed consent of all patients.
Tissue specimens
Paired fresh ESCC tissues and adjacent non-tumor tissues were obtained from 40 patients who underwent surgery for ESCC in Nanyang Central Hospital Affiliated Hospital of Henan University. After excision, the samples were immediately stored in a freezer at −80 °C. All specimens were confirmed pathologically as ESCC.
Cell culture and transfection
The human normal esophageal cell line HET1A and the ESCC cell lines TE1, EC109, TE13, and YES2 were from the American Type Culture Collection (ATCC, USA). All cells were maintained in RPMI 1640 Medium (Gibco) supplemented with 10% fetal bovine serum (Invitrogen) in an incubator at 37 °C containing 5% CO2. After appropriate time, the cells were transfected with Lipofectamine 2000 reagent (Invitrogen) in preparation for subsequent experiments. Full-length SNHG7 was amplified and inserted into a pcDNA3.1 vector to construct overexpression plasmid. Sequences of SNHG7 siRNAs were: sense, 5'-CGGAUUAUUUAGUCUUCAACA-3' and antisense, 5'-UUGAAGACUAAAUAAUCCGUU-3';
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from tissues and cells lines was isolated by using TRIzol reagent (Invitrogen), and subsequently reverse-transcribed into complementary DNA with a Prime-Script RT Reagent kit (Takara Biotechnology, Japan). Then, qRT-PCR reaction was conducted by using a SYBR Green kit (Applied Biosystems, USA). GAPDH and U6 snRNA were chosen to serve as internal controls for SNHG7 and miR-625, respectively. The 2-ΔΔCt method was applied to assess the relative gene expression.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay was carried out to investigate ESCC cell proliferation. Briefly, cells in the logarithmic growth phase were collected and resuspended to 2×104/mL. Then, 100 µL cells were seeded in 96-well plates and transfected with SNHG7-expressing plasmid and SNHG7 siRNA. At 24, 48, and 72 hours after transfection, 10 µL CCK8 solution (Beyotime Biotechnology) was added into each well. Cell were placed in an incubator for 3 hours, after which the absorbance was measured at wavelength of 450 nm.
5-Ethynyl-2'-deoxyuridine (EdU) experiment
For the EdU experiment, cells were routinely seeded in 6-well plates and transfected with SNHG7-expressing plasmid and SNHG7 siRNA. A Cell Light EdU Apollo 567 In Vitro Kit (Ribobio, China) was then used to measure the DNA replication activity. Images were collected with a fluorescence microscope to calculate the EdU positive rate.
Scratch assay
For the scratch assay, cells were collected after 48 hours of transfection, and 2×106 cells was added in a 6-well plate. The cells were cultured until reaching 90–100% confluence, at which point a linear scratch was made in the cell monolayer with a sterile 10-µL pipette tip. Cells were then incubated with FBS-free medium at 37 °C for 24 hours, and the speed of cell migration to the scratched area was observed. Images were collected with an optical microscope, and the scratch healing rate, representing the relative migration ability of the cells, was calculated with ImageJ software.
Transwell experiment
In Transwell assays, 1×105 transfected cells were re-suspended in 200 µL serum-free medium and inoculated into the upper chamber of a Transwell chamber (Corning, USA) coated with matrix gel. To the lower chamber, 600 µL of 10% FBS-containing medium was added. After 24-hour incubation at 37 °C with 5% CO2, the cells were subsequently fixed by 4% formaldehyde and stained by crystal violet reagent for 20 minutes. Cells invading through the chamber were counted with a microscope.
Dual-luciferase reporter gene assay
Plasmid of the wild-type (WT) reporter gene pmirGLO-SNHG7-wt containing the miR-625 binding site and the mutant plasmid pmirGLO-SNHG7-mut without the miR-625 binding site were constructed. The luciferase reporter plasmid and miR-625 mimics were co-transfected into cells by Lipofectamine 2000 reagent. At 48 hours after transfection, the relative luciferase activity was measured with a dual-luciferase reporter gene system (Promega).
Statistical analysis
All experiments in this study were independently repeated more than 3 times, and data were analyzed with the SPSS software (SPSS) and GraphPad Prism 7 (GraphPad, USA). Results were expressed as mean ± standard deviation. Student’s t test and one-way analysis of variance were applied to compare differences between group. P<0.05 was regarded as statistically significant.
Results
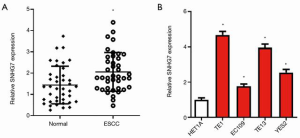
SNHG7 expression was increased in ESCC tissues and cell lines
To uncover the role of SNHG7 in ESCC, first, the expression level of SNHG7 was detected in 40 pairs of ESCC tissues and adjacent normal tissues. Compared with that in normal tissues, the level of SNHG7 in cancer tissues was significantly up-regulated (Figure 1A). We also used qRT-PCR to measure the expression of SNHG7 in HET1A and in ESCC cell lines. The data displayed that compared with that in HET1A cells, the level of SNHG7 in ESCC cells (TE1, EC109, TE13, and YES2) was significantly increased (Figure 1B). These findings indicated that SNHG7 expression was increased in ESCC tissues and cell lines.
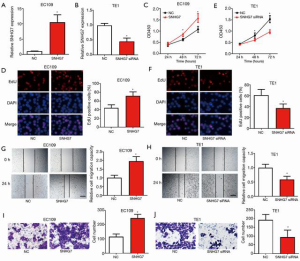
SNHG7 facilitated ESCC cell proliferation, migration, and invasion in vitro
To uncover the biological functions of SNHG7 in ESCC, we overexpressed or silenced SNHG7 in ESCC cells. As EC109 cells had the lowest SNHG7 level, they were transfected with SNHG7-expressing plasmid, and because TE1 cells exhibited the highest SNHG7 expression levels, they were transfected with SNHG7 small interfering RNA (siRNA). The transfection efficiency was verified by qRT-PCR (Figure 2A,B). After successful transfection, CCK8, EdU, scratch, and Transwell assays were carried out to study the influence of SNHG7 overexpression and knockdown on proliferation, migration, and invasion in vitro, respectively. The data of CCK8 and EdU experiments revealed that the proliferation of EC109 cells was obviously increased by SNHG7 overexpression (Figure 2C,D), while the proliferation ability of TE1 cells was significantly reduced after SNHG7 was down-regulated (Figure 2E,F). The results of the scratch experiment showed that EC109 cell migration was significantly increased by SNHG7 overexpression (Figure 2G), whereas the silencing of SNHG7 significantly reduced the migratory activity of TE1 cells (Figure 2H). The Transwell assay results demonstrated a significant increase in the invasion ability of EC109 cells after SNHG7 overexpression (Figure 2I), while a significant reduction was observed in the invasion ability of TE1 cells after SNHG7 silencing (Figure 2J). The above observations indicated that SNHG7 promoted ESCC cell proliferation, migration, and invasion in vitro.
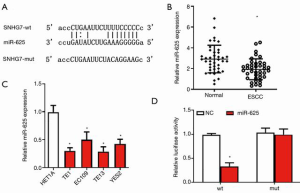
miR-625 is down-regulated and is a target gene of SNHG7 in ESCC
To further explore the downstream targets of SNHG7, the bioinformatics database (StarBase) was used to obtain potential target miRNAs of SNHG7. As shown in Figure 3A, the sequence of SNHG7 contains a conserved target site of miR-625. To examine the level of miR-625 in ESCC, we measured the level of miR-625 in ESCC tissues and cells by employing the qRT-PCR method. The results showed that compared with adjacent normal tissues, ESCC tissues exhibited significantly down-regulated miR-625 expression (Figure 3B). Further, the levels of miR-625 in ESCC cell lines (TE1, EC109, TE13, YES2) were significantly reduced compared with those in HET1A cells (Figure 3C). These findings indicated that miR-625 expression was decreased in ESCC. To confirm the direct interaction between SNHG7 and miR-625, a dual-luciferase reporter experiment was conducted. The data displayed that miR-625 mimics significantly inhibited the activity of luciferase plasmid with the wild-type SNHG7 sequence, whereas it played no effect on the mutant plasmid (Figure 3D). These results indicated that miR-625 is down-regulated in ESCC and can directly bind to SNHG7 as its target gene.
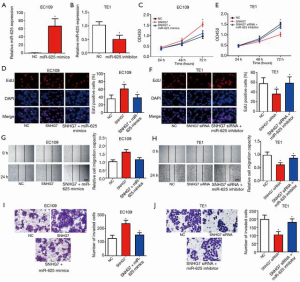
SNHG7 promotes the malignancy of ESCC by regulating miR-625
Studies have shown that miR-625 exerts an anti-tumor effect in ESCC. To further probe the role of the SNHG7/miR-625 signaling axis in ESCC cell proliferation, migration, and invasion, we conducted a rescue experiment. First, we successfully overexpressed and silenced miR-625 in ESCC cells (Figure 4A,B). Results showed that overexpressing SNHG7 facilitated the proliferation of EC109 cells, whereas overexpression of miR-625 partially eliminated the cancer-promoting effects of SNHG7 (Figure 4C,D). Similarly, silencing of SNHG7 inhibited EC109 cell proliferation, whereas silencing of miR-625 partially abolished the inhibitive effect of SNHG7 knockdown on cell proliferation (Figure 4E,F). SNHG7 overexpression promoted EC109 cell migration, whereas overexpression of miR-625 partially eliminated the migration-promoting effect of SNHG7 (Figure 4G). Silencing of SNHG7 also inhibited EC109 cell migration, while silencing of miR-625 partially abolished the inhibitive effect of SNHG7 on cell migration (Figure 4H). Furthermore, overexpression of SNHG7 promoted the invasion ability of EC109 cells, while overexpression of miR-625 partially abolished the pro-invasive effect of SNHG7 (Figure 4I). Similarly, the invasion ability of EC109 cells was inhibited when SNHG7 was silenced, and this effect was partially abolished by silencing of miR-625 (Figure 4J). These results indicated that SNHG7 and miR-625 have opposite biological functions in ESCC, with SNHG7 promoting the malignancy of ESCC cells via targeting the tumor suppressor miR-625.
Discussion
Non-coding RNAs (ncRNAs) have been a research hotspot in recent years, during which time miRNAs, lncRNAs, and circular RNAs have been confirmed to be important regulatory molecules in a variety of human cancers. Numerous reports have shown that lncRNAs are abnormally expressed in many cancers, and they play vital roles in regulating the pathological processes of cancer, such as cell proliferation, metabolism, epithelial-mesenchymal transition, migration, and invasion (13,14). Small nucleolar RNA host genes (SNHGs), for instance, play extremely important regulatory roles in ESCC. Specifically, lncRNA-SNHG6 can inhibit ESCC cell apoptosis by targeting miR-101-3p to regulate Enhancer of zeste homolog 2 (EZH2) (15). Furthermore, SNHG5 can inhibit the epithelial-mesenchymal transition process of ESCC cells through ubiquitination and degradation of metastasis-associated protein 2 (MTA2) (16). As for SNHG7, there is evidence that it is correlated to the occurrence, development, as well as the poor prognosis of various cancers, including renal cell carcinoma, hepatocellular carcinoma, and lung cancer (17). Yet, the expression, functions, and mechanism of SNHG7 in ESCC are still unknown. Therefore, the pioneering study has become the first to explore SNHG7 in ESCC.
Our results revealed that the expression of SNHG7 in ESCC tissues and cell lines is obviously higher than that in normal tissues and cell lines. At the same time, we confirmed through in vitro assays that SNHG7 overexpression is able to significantly facilitate ESCC cells proliferation, migration and invasion, while silencing of SNHG7 can inhibit these activities in ESCC cells. These results suggest that SNHG7 plays a carcinogenic role in ESCC. In almost all tumor-related studies involving SNHG7, this lncRNA has been confirmed to be up-regulated in tumors and to promote the malignant progression of cancer cells by enhancing their proliferation, migration, invasion, drug resistance, survival, and other biological processes in vivo and in vitro (17). Our study revealed that SNHG7 can obviously accelerate ESCC cells proliferation, migration, and invasion, and play an oncogenic role to promote malignant tumor progression. Therefore, SNHG7 may be an oncogene in various tumors and is expected to become a potential target for cancer therapy.
Studies have shown that many lncRNAs can interact with miRNAs as competitive endogenous RNAs (ceRNAs), thereby inhibiting the function of these miRNAs and regulating the expression of their target genes. As 1 example, SNHG7 may play an oncogenic role by targeting multiple tumor-suppressor miRNAs. In small cell lung cancer, for instance, SNHG7 acts as a sponge of miR-181a-5p, and it also participates in the processes of non-small cell lung cancer through its regulation of a miR-181a-5p/E2F7 signaling (18). In hepatocellular carcinoma, SNHG7 directly targets miR-9-5p, thereby up-regulating the expression of CNNM1 and promoting the occurrence of this tumor (12). LncRNAs and miRNAs are interrelated with each other to form a lncRNA-mediated ceRNA regulatory network. Increasing evidences show that this network of ceRNAs is closely related to tumorigenesis (19). To date, studies have uncovered numerous lncRNA-mediated ceRNA regulatory networks in ESCC (20). In the present study, we found that the SNHG7 sequence contained a conserved target of miR-625, and at the same time, we confirmed that the expression level of miR-625 in ESCC tissues and cells was decreased. The dual-luciferase reporter gene experiment results showed that miR-625 can directly bind to the SNHG7 sequence and serves as the target gene of SNHG7.
The expression of miR-625 in ESCC tissues has been reported to be significantly lower than that in adjacent normal tissues. Low miR-625 expression is closely related to lymph node metastasis, distant metastasis, tumor differentiation, tumor-node-metastasis staging, and a low 5-year overall survival rate. Li et al. described low miR-625 expression to be an independent predictive factor of ESCC prognosis (21), and Wang et al. reported that it was able to facilitated EC cells proliferation and invasion, and may regulate the biological processes of EC by controlling the expression of Sox2 (22). Therefore, these studies suggest that miR-625 is likely to serve as a tumor-suppressor gene in ESCC, consistenting with the results of the present study and confirms the idea that SNHG7 promotes ESCC malignancy by suppressing miR-625. Finally, we verified that SNHG7 regulates the proliferation, migration and invasion of ESCC cells by mediating miR-625 to establish an SNHG7/miR-625 regulatory network, which further substantiates the regulatory network of SNHG7 and further supplements the pathogenesis of ESCC.
However, the current research did not include animal experiments, meaning the roles of SNHG7 on ESCC tumor growth and metastasis in vivo could not be definitively explained, which is the main limitation of this study. Therefore, the biological effects of SNHG7 and its corresponding molecular mechanism will be further explored in future studies involving in vivo experiments.
Conclusions
In summary, we showed that SNHG7 is up-regulated in ESCC tissues and cell lines, whereas the expression levels of miR-625 are low. SNHG7 facilitates the proliferation, migration, and invasion of ESCC cells by interacting with miR-625. This study has revealed the regulatory role of the SNHG7/miR-625 axis in ESCC, providing a new potential target for the treatment of ESCC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-21-147
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-21-147
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (Available at http://dx.doi.org/10.21037/jgo-21-147). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Nanyang Central Hospital Affiliated Hospital of Henan University (No. 20160075), and the specimens were obtained with the informed consent of all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yang YM, Hong P, Xu WW, et al. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther 2020;5:229. [Crossref] [PubMed]
- Yoshida N, Eto K, Kurashige J, et al. Comprehensive Analysis of Multiple Primary Cancers in Patients with Esophageal Squamous Cell Carcinoma Undergoing Esophagectomy. Ann Surg 2020; Epub ahead of print. [Crossref] [PubMed]
- Wen J, Wang G, Xie X, et al. Prognostic Value of a Four-miRNA Signature in Patients With Lymph Node Positive Locoregional Esophageal Squamous Cell Carcinoma Undergoing Complete Surgical Resection. Ann Surg 2021;273:523-31. [Crossref] [PubMed]
- Shin DW, Kim HK, Cho J, et al. Conditional Survival of Patients Who Underwent Curative Resection for Esophageal Squamous Cell Carcinoma. [Epub ahead of print]. doi:
10.1097/SLA.0000000000004473 . - Hu Q, Egranov SD, Lin C, et al. Long noncoding RNA loss in immune suppression in cancer. Pharmacol Ther 2020;213:107591 [Crossref] [PubMed]
- Aprile M, Katopodi V, Leucci E, et al. LncRNAs in Cancer: From garbage to Junk. Cancers (Basel) 2020;12:3220. [Crossref] [PubMed]
- Feng B, Wang G, Liang X, et al. LncRNA FAM83H-AS1 promotes oesophageal squamous cell carcinoma progression via miR-10a-5p/Girdin axis. J Cell Mol Med 2020;24:8962-76. [Crossref] [PubMed]
- Sun K, Zhang G. Long noncoding RNA CASC2 suppresses esophageal squamous cell carcinoma progression by increasing SOCS1 expression. Cell Biosci 2019;9:90. [Crossref] [PubMed]
- Li L, Ye D, Liu L, et al. Long Noncoding RNA SNHG7 Accelerates Proliferation, Migration and Invasion of Non-Small Cell Lung Cancer Cells by Suppressing miR-181a-5p Through AKT/mTOR Signaling Pathway. Cancer Manag Res 2020;12:8303-12. [Crossref] [PubMed]
- Chen Y, Yuan S, Ning T, et al. SNHG7 Facilitates Glioblastoma Progression by Functioning as a Molecular Sponge for MicroRNA-449b-5p and Thereby Increasing MYCN Expression. Technol Cancer Res Treat 2020;19:1533033820945802 [Crossref] [PubMed]
- Boone DN, Warburton A, Som S, et al. SNHG7 is a lncRNA oncogene controlled by Insulin-like Growth Factor signaling through a negative feedback loop to tightly regulate proliferation. Sci Rep 2020;10:8583. [Crossref] [PubMed]
- Xie Y, Wang Y, Gong R, et al. SNHG7 Facilitates Hepatocellular Carcinoma Occurrence by Sequestering miR-9-5p to Upregulate CNNM1 Expression. Cancer Biother Radiopharm 2020;35:731-40. [Crossref] [PubMed]
- Feng Q, Zhang H, Yao D, et al. Emerging Role of Non-Coding RNAs in Esophageal Squamous Cell Carcinoma. Int J Mol Sci 2019;21:258. [Crossref] [PubMed]
- Zhang Y, Liu Q, Liao Q. Long noncoding RNA: a dazzling dancer in tumor immune microenvironment. J Exp Clin Cancer Res 2020;39:231. [Crossref] [PubMed]
- Wang J, Yang X, Li R, et al. LncRNA SNHG6 Inhibits Apoptosis by Regulating EZH2 Expression via the Sponging of MiR-101-3p in Esophageal Squamous-Cell Carcinoma. Onco Targets Ther 2020;13:11411-20. [Crossref] [PubMed]
- Wei S, Sun S, Zhou X, et al. SNHG5 inhibits the progression of EMT through the ubiquitin-degradation of MTA2 in oesophageal cancer. Carcinogenesis 2021;42:315-26. [Crossref] [PubMed]
- Zhou Y, Tian B, Tang J, et al. SNHG7: A novel vital oncogenic lncRNA in human cancers. Biomed Pharmacother 2020;124:109921 [Crossref] [PubMed]
- Wang L, Zhang L, Wang L. SNHG7 Contributes to the Progression of Non-Small-Cell Lung Cancer via the SNHG7/miR-181a-5p/E2F7 Axis. Cancer Manag Res 2020;12:3211-22. [Crossref] [PubMed]
- Xu G, Xu WY, Xiao Y, et al. The emerging roles of non-coding competing endogenous RNA in hepatocellular carcinoma. Cancer Cell Int 2020;20:496. [Crossref] [PubMed]
- Wang QY, Peng L, Chen Y, et al. Characterization of super-enhancer-associated functional lncRNAs acting as ceRNAs in ESCC. Mol Oncol 2020;14:2203-30. [Crossref] [PubMed]
- Li C, Li DC, Che SS, et al. The decreased expression of miR-625 predicts poor prognosis of esophageal squamous cell carcinoma. Int J Clin Exp Med 2015;8:9560-4. [PubMed]
- Wang Z, Qiao Q, Chen M, et al. miR-625 down-regulation promotes proliferation and invasion in esophageal cancer by targeting Sox2. FEBS Lett 2014;588:915-21. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


