Molecular testing to optimize therapeutic decision making in advanced colorectal cancer
Introduction
Despite the adoption of widespread screening efforts, a significant fraction of the 136,830 patients diagnosed with colon or rectal cancer will ultimately succumb to advanced disease (1). Indeed, colorectal cancer (CRC) is the second leading cause of cancer death in the United States, accounting for an estimated 50,310 deaths in 2014 (1). Over the past several decades, there have been a number of changes in the treatment options for patients with advanced CRC that include the incorporation of new drugs as well as the recognition of the benefit of surgical resection for selected patients with metastatic disease.
The first drug to demonstrate activity for the treatment of advanced CRC was 5-fluorouracil (5-FU). Initial efforts sought to optimize the dose and schedule of this agent while demonstrating the benefit of the addition of leucovorin. Over the past couple of decades, two additional cytotoxic agents have been added to the therapeutic arsenal for the treatment of advanced CRC. Irinotecan is an intravenous camptothecin analog that was initially developed for advanced CRC that was refractory to leucovorin-modulated 5-FU and is now incorporated into multiple lines of therapy (2,3). Oxaliplatin is a platinum derivative that has minimal single-agent activity in advanced CRC. However, the drug has substantial activity when combined with a fluoropyrimidine (4).
In recent years, two classes of biologic agents have been developed to advance outcomes in this disease by targeting vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) signaling respectively. Bevacizumab is a monoclonal antibody that binds to circulating VEGF isoform A with activity in multiple lines of treatment in advanced CRC (5). Ziv-aflibercept is a related agent that binds to multiple VEGF isoforms, and is now approved for use in the second-line in combination with irinotecan-based chemotherapy (6). Regorafenib is an oral multi-kinase inhibitor with anti-VEGF as well as potential anti-proliferative effect (7). While not solely an anti-VEGF agent, it perhaps can best be categorized as an extension of the anti-angiogenic paradigm in CRC. Cetuximab is a recombinant mouse/human monoclonal antibody that binds to the EGFR and was the first EGFR-targeting agent to demonstrate activity in irinotecan- and oxaliplatin-refractory CRC (8). Its use has now been expanded to multiple lines of therapy (9-11). Panitumumab is a very similar human monoclonal antibody targeting EGFR that has comparable activity to cetuximab as a single agent and in combination with chemotherapy (12,13).
While the application of each of these new treatments for advanced CRC has pushed the median survival to approximately 30 months or greater in recent clinical trials (14,15), each individual agent is not universally effective and is associated with nontrivial toxicity. Therefore, significant recent efforts have been made to understand both factors associated with survival regardless of therapy (prognostic biomarkers) and factors that identify patients more or less likely to benefit from a specific intervention (predictive biomarkers). In this review, we will discuss the current status of molecular biomarkers to select chemotherapy, anti-EGFR agents, and anti-VEGF agents in advanced CRC focusing on clinical studies and with a look forward to emerging and future developments.
Molecular tests to individualize cytotoxic chemotherapy
Uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1)
UGT1A1 is the key enzyme for inactivation of the active metabolite of irinotecan, SN-38 through glucuronidation. The UGT1A1*28 polymorphism is characterized by an extra TA repeat in the gene’s promoter region, and is associated with reduced protein expression and therefore reduced glucuronidation. The presence of two UGT1A1*28 alleles underlies Gilbert’s syndrome. Initial studies demonstrated that toxicity, particularly hematologic toxicity, was significantly increased in patients with one or two alleles of UGT1A1*28 (16). This resulted in its notation in the prescribing information; however, management strategies for patients homozygous or heterozygous for this allele have not been standardized. In a study of 250 Caucasian patients with advanced CRC treated with the FOLFIRI (5-FU, leucovorin, irinotecan) regimen, including 22 (8.8%) homozygous and 114 (45.6%) heterozygous for UGT1A1*28, this marker was associated with an increased risk of hematologic toxicity in the first cycle but without statistically significant effect in subsequent cycles (17). A recent phase I study used flat doses of irinotecan to establish the maximum tolerated dose according to UGT1A1 genotype for an every 3-week regimen (18). In addition to the established association of UGT1A1*28 polymorphisms with irinotecan-associated toxicity, it has also been hypothesized that the reduced rate of elimination may actually be a predictive marker for response. Indeed, in the above-mentioned study of patients with CRC treated with FOLFIRI, response was increased in the UGT1A1*28 homozygous group (17). Nonetheless, this did not translate into a statistically significant increase in survival. While it is clear that genetic heterogeneity at UGT1A1 is associated with altered metabolism of SN-38, other drug-metabolizing enzymes influence the final toxicity and efficacy outcomes. The optimal management of irinotecan based on UGT1A1 status therefore remains incompletely defined (19).
Excision repair and cross-complementation group 1 (ERCC1)
The ERCC1 protein is a key component of the nucleotide excision repair complex, which is a primary method of repair of platinum-DNA adducts. Therefore, it has been hypothesized that low ERCC1 expression may be associated with sensitivity to oxaliplatin-based chemotherapy in cancer (20). An initial study of 50 patients treated with an oxaliplatin and 5-FU combination regimen suggested that a group of patients with higher RNA expression of ERCC1 had a 4.8-fold higher risk of death (95% CI, 2.09-15.88) compared to those with lower expression (21). In a 91-patient phase I study of escalating doses of capecitabine with oxaliplatin, ERCC1 RNA expression in a metastatic site was associated with time to treatment failure (22). Kim et al. studied expression of ERCC1 by immunohistochemistry (IHC) in 70 patients with advanced CRC treated with an oxaliplatin-containing regimen (22). Survival was longer in those who did not have detectable ERCC1 expression according to this method (P=0.0474). However, this was not confirmed in a much larger analysis (n=1,197) of the MRC FOCUS trial, which failed to confirm any predictive benefit of ERCC1 IHC (23). Questions have been raised about the specificity of the antibody used to assess ERCC1 and about the concordance of ERCC1 expression in the primary and metastatic tumor tissue (24,25). Thus, the optimal method of assessment of assessment of ERCC1 status is not yet clearly defined, as different methods may give different results (26).
A large number of studies have evaluated the predictive utility of a polymorphism in ERCC1 (ERCC1-118 C>T) to predict outcomes of patients treated with oxaliplatin. While several studies have suggested that the T containing genotype is associated with better response and/or survival (27,28), several others have come to the exact opposite conclusion (29-31), and indeed further studies suggest no association (32,33). A potential explanation for this discrepant data is that the polymorphism may have different implications in a Caucasian versus Asian population (34,35). Given the limited and contradictory clinical data, measuring ERCC1 expression or gene polymorphisms by any method cannot be recommended for use to select oxaliplatin-based treatment at this time.
Microsatellite instability (MSI)
Approximately 15% of CRCs have evidence of defective DNA mismatch repair manifested as MSI. While some of these tumors are a result of hereditary defects in DNA mismatch repair enzymes (hereditary non-polyposis CRC), others are sporadic and the result of promoter silencing of the DNA mismatch repair enzyme MLH1 (36). Current data suggest that defective DNA mismatch repair confers an improved prognosis in most stages of CRC, but there is also evidence that these tumors are relatively resistant to 5-FU monotherapy in the adjuvant setting (37). The utility of MSI testing to direct treatment in advanced CRC is not established. A summary of candidate markers is provided in Table 1.
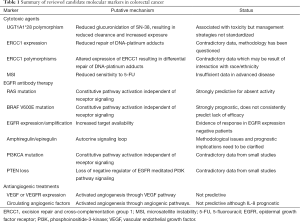
Full table
Molecular tests to individualize anti-EGFR therapy
KRAS and NRAS
The EGFR signaling pathway has been targeted in a number of different malignancies. Upon binding to ligands such as EGF, amphiregulin, or epiregulin, EGFR results in an intracellular signal that is transduced through several intracellular signaling pathways. A dominant pathway involves the activation of the G-protein intermediate RAS, and subsequent signaling through BRAF, MEK, and ERK (the MAP kinase pathway). Mutations in the RAS family of proto-oncogenes (KRAS, NRAS, HRAS) result in constitutive activation of MAP kinase pathway signaling that is independent of activation of receptor tyrosine kinases such as EGFR. Since the currently available EGFR antibodies cetuximab and panitumumab bind to the extracellular domain of EGFR to result in receptor internalization and block signaling, it has been hypothesized that mutations in RAS render the activity of EGFR antibodies irrelevant.
The most common RAS mutations in colon cancer occur in codons 12 and 13 of KRAS, and are present in 35-45% of cases. An initial study of 30 patients confirmed preclinical data that tumors harboring this mutation would be resistant to EGFR antibody therapy (38). These findings were subsequently confirmed in prospective retrospective analyses of large clinical trials that randomized patients to treatment with or without EGFR antibodies. For example, in the CRYSTAL trial that compared initial treatment with the combination of leucovorin, 5-FU, and irinotecan (FOLFIRI) to the same treatment with cetuximab, there was a significant interaction between KRAS mutation status and survival outcome (9). Many other similar studies and a meta-analysis came to similar conclusions (12,39-41). Therefore, KRAS mutation testing has been integrated into guideline-based management of advanced CRC (42).
Multiple questions have been raised by these data, perhaps most prominently is whether all mutations in RAS behave similarly. Initially, it was questioned whether codon 13 mutations were equivalent to codon 12 mutations in predicting the lack of efficacy of EGFR antibodies. Several retrospective studies suggested that the predictive effect of KRAS codon 13 mutation was incomplete (43,44); nonetheless, targeted studies have not suggested that benefit in this population is worthy of changing the paradigm (45).
Subsequently, other mutations in KRAS and similar mutations in NRAS have been evaluated as potential predictors of lack of response to EGFR antibodies in CRC (46). The other mutations frequently encountered include mutations in codons 59, 61, 117, and 146. They are generally activating in cell line models (47). Recent secondary analysis of several key studies of EGFR antibodies in the treatment of advanced CRC have demonstrated no trend for benefit for the addition of EGFR antibodies of patients to patients whose tumors these further RAS mutations (which we and others refer to as expanded RAS testing, see Table 2) (10,48-52). While no single study is statistically powered to evaluate the treatment by biomarker interaction for each of these additional mutations individually, this extended analysis has generally been accepted as standard prior to the prescription of an anti-EGFR agent (53). Moreover, with the application of next generation sequencing technology to simultaneous assessment of multiple candidate genetic markers, further refinement of this algorithm can be expected (54).
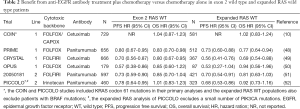
Full table
BRAF
Using analogous reasoning to that described for RAS mutations, the BRAF V600E mutation, immediately downstream of RAS, has been evaluated as a negative predictive marker for the efficacy of EGFR antibodies. However, establishing the BRAF V600E mutation as a negative predictive marker has been challenging due to its lower prevalence (approximately 5-10%) (39-41,55). Additionally, an activating mutation in BRAF conveys a strong prognostic significance, with mutated tumors conferring a poor prognosis (10). Most studies do not clearly demonstrate a negative predictive benefit for BRAF for the selection of EGFR antibodies (48). Nonetheless, BRAF mutation testing has been recommended due to its strong implications on prognosis and due to the availability of BRAF mutation targeted clinical trials (56). Indeed, we use this information to guide the intensity of therapy and surveillance for progression of disease on treatment.
EGFR expression and amplification
During the initial development of EGFR antibodies in metastatic CRC, it was predicted that EGFR protein expression would be required for any activity. Therefore, only EGFR-expressing CRC was allowed on the initial clinical trials (57). However, subsequent analyses did not demonstrate a correlation with EGFR expression and response (58). Response to an EGFR antibody was observed in tumors that did not have immunohistochemically detectable expression of EGFR (59). Therefore, expression of EGFR has been abandoned as a predictive marker for the benefit of EGFR antibody therapy CRC. Similarly, EGFR copy number has been described as a poor candidate for development as a predictive marker of the efficacy of EGFR antibody therapy (60), despite at least one small study suggesting a provocative interaction between copy number and outcome (61).
EGFR ligand expression
Intratumoral expression of the EGFR ligands amphiregulin and epiregulin may reflect activation of the EGFR pathway through an autocrine signaling loop. An initial study using tissue obtained from pretreatment tumor biopsy mRNA on a single agent cetuximab study suggested that intratumor expression was strongly associated with disease control and progression free survival (62). A follow up analysis of tissue obtained on a study combining cetuximab and irinotecan demonstrated concordant results, and further demonstrated that the effect was only relevant in tumors with absent mutation in KRAS (63). However, ligand expression may have prognostic implications that dilute the predictive utility and the optimal method of measurement is not yet standardized as concordance between tumor sites is modest (64,65). Thus, while the measurement of amphiregulin or epiregulin expression cannot be recommended to select EGFR antibody use in RAS wild type patients, they remain strong candidates for further development in ongoing studies of EGFR antibody therapy in CRC.
PI3K pathway alterations
In addition to the RAS pathway, EGFR signaling is mediated through a distinct pathway that involves phosphoinositide-3-kinase (PI3K), AKT and PTEN. Similar to the rationale for how RAS pathway mutations impact upstream EGFR-directed therapy, mutations in the PI3K pathway have been evaluated as a predictive marker for the efficacy of EGFR antibody therapy.
The α-catalytic domain of PI3K is encoded by the PIK3CA gene and is mutated in 10-20% of CRCs. Mutation in PIK3CA is not mutually exclusive with RAS mutations. The majority of mutations in PIK3CA occur in either exon 9 (the helical domain) or exon 20 (the kinase domain). Initial studies came to conflicting conclusions regarding the potential utility of PIK3CA mutations in predicting resistance to EGFR-targeted therapy (66,67). In a large study of 773 patients with evaluable specimens treated with EGFR antibody therapy, PIK3CA mutations were associated with a lower response rate in KRAS wild-type patients (17.7% vs. 37.7%; OR 0.35; 95% CI, 0.13-0.83; P=0.015) but not with reduced PFS or OS (40). Interestingly, the effect seemed to be confined to exon 20 mutations where response, PFS and OS were all statistically reduced compare to wild type patients. Nonetheless, exon 20 mutations are relatively rare (only 9 without KRAS mutation were analyzed in the study) and further studies are needed to confirm these findings and establish the magnitude of the effect. Indeed, a subsequent study on a subset of patients from the original cetuximab monotherapy phase III study did not find any effect of PIK3CA mutations as a predictive marker for benefit (41).
Phosphatase and tensin homologue deleted on chromosome ten (PTEN) is a tumor suppressor gene which functions as a negative regulator of PI3K activation by EGFR. Loss of PTEN function results in sustained activation of PI3K effector proteins and has been reported to occur in the range of 20-40% of CRC (68,69). In small retrospective series of 27 patients with CRC who received cetuximab as part of their therapy, 10 of 16 patients with intact PTEN had objective response in contrast to none of the 11 patients with loss of PTEN (68). Interpretation of this study is limited by both the small numbers but also presence of KRAS mutations in 37% of the cases. A larger but also retrospective study found loss of PTEN in 20% of 111 cases of wild type KRAS CRC (69). In contrast to the prior study, loss of PTEN was not associated with response or progression-free survival although overall survival was shorter.
Molecular tests to individualize anti-VEGF therapy
A number of markers have been explored as potential predictors for the benefit of anti-VEGF strategies in advanced CRC. Initial studies hypothesized that altered intratumoral expression of VEGF would predict responsiveness to bevacizumab. However, examination of VEGF expression by in situ hybridization and immunohistochemistry did not correlate with outcome in a subset analysis of the pivotal AVF2107g study (70). Similarly, immunohistochemical analysis from the NO166966 study of XELOX and FOLFOX with or without bevacizumab did not demonstrate an association of VEGF, VEGFR1, or VEGFR2 on tumor cells or stroma with progression free survival (71).
Alternatively, it was proposed that baseline or pharmacodynamic changes in circulating angiogenic factors such as VEGF-A could predict efficacy of bevacizumab. This was initially piloted in a phase II study of bevacizumab in rectal cancer that demonstrated no evidence for a predictive benefit of baseline plasma VEGF (72). Nonetheless, provocative data were generated that associated baseline soluble VEGFR1 and increases (>2-fold) in plasma PlGF with improved treatment effect. In the HORIZON studies testing the anti-VEGF kinase inhibitor cediranib, high circulating VEGF was associated with worse outcome. However, neither clear predictive effect of baseline VEGF nor soluble VEGFR2 was observed for the efficacy of cediranib versus placebo (HORIZON II) or cediranib versus bevacizumab (HORIZON III) (73). In a phase II study of FOLFIRI plus bevacizumab for patients with previously untreated advanced CRC, Kopetz et al. analyzed a panel of 37 baseline and on-treatment plasma angiogenic factors for association with PFS (74). Notably, both baseline VEGF and VEGFR2 were not associated with outcome. However, high baseline interleukin-8 levels were associated with a shorter PFS (11 vs. 15.1 months; P=0.03). Moreover, elevations in bFGF, PlGF, and HGF were observed in subsets of patients before radiographic evidence of disease progression. Despite these interesting data, there is no marker available to select for benefit of anti-angiogenic therapies at this time in advanced CRC or elsewhere (75,76).
Expression profiling
Recognizing that single molecular events only partly define the molecular architecture of CRC, a number of investigators have explored gene expression profiling as a mechanism to characterize subsets of CRCs that are likely to respond to a given intervention. For example, a relatively small study used an institutional training set to identify 7 genes from a Affymetrix U133 Plus 2.0 chip that were differentially expressed in patients who responded to 5-FU-based chemotherapy (77). After converting the signature to a RT-qPCR assay, the authors demonstrated that patients with a favorable predictive signature had a significantly greater response rate (58% vs. 13%, P=0.024), improved PFS (61% vs. 13% at 1 year, HR =0.32, P=0.009), and improved OS (32 vs. 16 months, HR =0.21, P=0.003). Such data are provocative and clearly larger studies are warranted.
Conclusions
It is an exciting time to be treating advanced CRC as the development of multiple new treatment strategies has pushed the median survival to the range of 30 months. Nonetheless, the availability of selection markers to optimize therapy on an individual patient basis has not kept up with the pace of drug development. It has been established that UGT1A1*28 polymorphism is associated with irinotecan toxicity, although the optimal management strategy for this marker has not been identified. Performance of a test for mutations in exon 2 of KRAS has now been expanded to other sites in exons 3 and 4 of KRAS and analogous sites in NRAS, and is essential for any patient being considered for an EGFR targeted antibody. A mutation in BRAF (V600E) is associated with a poor prognosis, which may drive alternative treatment strategies for individual patients. Unfortunately, no established predictive biomarker for anti-VEGF therapy yet exists. Nonetheless, the application of new technologies and robust study designs to biomarker discovery and validation efforts is likely to expand the library of available molecular tests to optimize care for patients with advanced CRC in the near future.
Acknowledgements
Funding: TJ Semrad and EJ Kim are supported by the National Cancer Institute of the National Institutes of Health under award number K12CA138464.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [PubMed]
- Cunningham D, Glimelius B. A phase III study of irinotecan (CPT-11) versus best supportive care in patients with metastatic colorectal cancer who have failed 5-fluorouracil therapy. V301 Study Group. Semin Oncol 1999;26:6-12. [PubMed]
- Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet 2000;355:1041-7. [PubMed]
- André T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med 2004;350:2343-51. [PubMed]
- Bennouna J, Sastre J, Arnold D, et al. Continuation of bevacizumab after first progression in metastatic colorectal cancer (ML18147): a randomised phase 3 trial. Lancet Oncol 2013;14:29-37. [PubMed]
- Van Cutsem E, Tabernero J, Lakomy R, et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. J Clin Oncol 2012;30:3499-506. [PubMed]
- Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet 2013;381:303-12. [PubMed]
- Jonker DJ, O’Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040-8. [PubMed]
- Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 2011;29:2011-9. [PubMed]
- Maughan TS, Adams RA, Smith CG, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet 2011;377:2103-14. [PubMed]
- Bokemeyer C, Bondarenko I, Makhson A, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 2009;27:663-71. [PubMed]
- Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 2010;28:4697-705. [PubMed]
- Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 2007;25:1658-64. [PubMed]
- Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol 2014;15:1065-75. [PubMed]
- Venook AP, Niedzwiecki D, Lenz HJ, et al. CALGB/SWOG 80405: Phase III trial of irinotecan/5-FU/leucovorin (FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS wild-type (wt) untreated metastatic adenocarcinoma of the colon or rectum (MCRC). J Clin Oncol 2014;32:abstr LBA3.
- Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J 2002;2:43-7. [PubMed]
- Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol 2006;24:3061-8. [PubMed]
- Innocenti F, Schilsky RL, Ramirez J, et al. Dose-finding and pharmacokinetic study to optimize the dosing of irinotecan according to the UGT1A1 genotype of patients with cancer. J Clin Oncol 2014;32:2328-34. [PubMed]
- Fujita K, Sparreboom A. Pharmacogenetics of irinotecan disposition and toxicity: a review. Curr Clin Pharmacol 2010;5:209-17. [PubMed]
- Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res 2008;14:1291-5. [PubMed]
- Shirota Y, Stoehlmacher J, Brabender J, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol 2001;19:4298-304. [PubMed]
- Kim SH, Kwon HC, Oh SY, et al. Prognostic value of ERCC1, thymidylate synthase, and glutathione S-transferase pi for 5-FU/oxaliplatin chemotherapy in advanced colorectal cancer. Am J Clin Oncol 2009;32:38-43. [PubMed]
- Braun MS, Richman SD, Quirke P, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol 2008;26:2690-8. [PubMed]
- Ma D, Baruch D, Shu Y, et al. Using protein microarray technology to screen anti-ERCC1 monoclonal antibodies for specificity and applications in pathology. BMC Biotechnol 2012;12:88. [PubMed]
- Kobayashi H, Sugihara K, Uetake H, et al. Messenger RNA expression of TS and ERCC1 in colorectal cancer and matched liver metastasis. Int J Oncol 2008;33:1257-62. [PubMed]
- Bohanes P, Labonte MJ, Lenz HJ. A review of excision repair cross-complementation group 1 in colorectal cancer. Clin Colorectal Cancer 2011;10:157-64. [PubMed]
- Viguier J, Boige V, Miquel C, et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res 2005;11:6212-7. [PubMed]
- Paré L, Marcuello E, Altes A, et al. Pharmacogenetic prediction of clinical outcome in advanced colorectal cancer patients receiving oxaliplatin/5-fluorouracil as first-line chemotherapy. Br J Cancer 2008;99:1050-5. [PubMed]
- Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol 2007;25:1247-54. [PubMed]
- Stoehlmacher J, Park DJ, Zhang W, et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer 2004;91:344-54. [PubMed]
- Chua W, Goldstein D, Lee CK, et al. Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer 2009;101:998-1004. [PubMed]
- Etienne-Grimaldi MC, Milano G, Maindrault-Goebel F, et al. Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br J Clin Pharmacol 2010;69:58-66. [PubMed]
- Spindler KL, Andersen RF, Jensen LH, et al. EGF61A>G polymorphism as predictive marker of clinical outcome to first-line capecitabine and oxaliplatin in metastatic colorectal cancer. Ann Oncol 2010;21:535-9. [PubMed]
- Qian YY, Liu XY, Wu Q, et al. The ERCC1 C118T polymorphism predicts clinical outcomes of colorectal cancer patients receiving oxaliplatin-based chemotherapy: a meta-analysis based on 22 studies. Asian Pac J Cancer Prev 2014;15:8383-90. [PubMed]
- Lu X, Xiao S, Jin C, et al. ERCC1 and XPD/ERCC2 polymorphisms’ predictive value of oxaliplatin-based chemotherapies in advanced colorectal cancer has an ethnic discrepancy: a meta-analysis. J Clin Lab Anal 2012;26:10-5. [PubMed]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003;348:919-32. [PubMed]
- Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol 2010;28:3219-26. [PubMed]
- Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66:3992-5. [PubMed]
- Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 2011;22:1535-46. [PubMed]
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [PubMed]
- Karapetis CS, Jonker D, Daneshmand M, et al. PIK3CA, BRAF, and PTEN status and benefit from cetuximab in the treatment of advanced colorectal cancer--results from NCIC CTG/AGITG CO.17. Clin Cancer Res 2014;20:744-53. [PubMed]
- Benson AB 3rd, Venook AP, Bekaii-Saab T, et al. Colon cancer, version 3.2014. J Natl Compr Canc Netw 2014;12:1028-59. [PubMed]
- De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 2010;304:1812-20. [PubMed]
- Tejpar S, Celik I, Schlichting M, et al. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol 2012;30:3570-7. [PubMed]
- Schirripa M, Lonardi S, Cremolini C, et al, editors. Phase II Study of Single Agent Cetuximab in KRAS G13D Mutant Metastatic Colorectal Cancer (MCRC). Oxford, England: Oxford Univ Press, 2014.
- Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res 2010;70:5901-11. [PubMed]
- Smith G, Bounds R, Wolf H, et al. Activating K-Ras mutations outwith 'hotspot' codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer 2010;102:693-703. [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [PubMed]
- Van Cutsem E, Lenz HJ, Kohne CH, et al. Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J Clin Oncol 2015;33:692-700. [PubMed]
- Bokemeyer C, Kohne CH, Ciardiello F, et al. Treatment outcome according to tumor RAS mutation status in OPUS study patients with metastatic colorectal cancer (mCRC) randomized to FOLFOX4 with/without cetuximab. J Clin Oncol 2014;32:abstr 3505.
- Peeters M, Oliner K, Price T, et al. Updated analysis of KRAS/NRAS and BRAF mutations in study 20050181 of panitumumab (pmab) plus FOLFIRI for second-line treatment (tx) of metastatic colorectal cancer (mCRC). J Clin Oncol 2014;35:abstr 3568.
- Seymour MT, Brown SR, Middleton G, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol 2013;14:749-59. [PubMed]
- Sorich MJ, Wiese MD, Rowland A, et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol 2015;26:13-21. [PubMed]
- Peeters M, Oliner KS, Parker A, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res 2013;19:1902-12. [PubMed]
- De Roock W, De Vriendt V, Normanno N, et al. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol 2011;12:594-603. [PubMed]
- Atreya CE, Corcoran RB, Kopetz S. Expanded RAS: Refining the Patient Population. J Clin Oncol 2015;33:682-5. [PubMed]
- Saltz LB, Meropol NJ, Loehrer PJ Sr, et al. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol 2004;22:1201-8. [PubMed]
- Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004;351:337-45. [PubMed]
- Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005;23:1803-10. [PubMed]
- Italiano A, Follana P, Caroli FX, et al. Cetuximab shows activity in colorectal cancer patients with tumors for which FISH analysis does not detect an increase in EGFR gene copy number. Ann Surg Oncol 2008;15:649-54. [PubMed]
- Scartozzi M, Bearzi I, Mandolesi A, et al. Epidermal Growth Factor Receptor (EGFR) gene copy number (GCN) correlates with clinical activity of irinotecan-cetuximab in K-RAS wild-type colorectal cancer: a fluorescence in situ (FISH) and chromogenic in situ hybridization (CISH) analysis. BMC Cancer 2009;9:303. [PubMed]
- Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 2007;25:3230-7. [PubMed]
- Jacobs B, De Roock W, Piessevaux H, et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol 2009;27:5068-74. [PubMed]
- Li XD, Miao SY, Wang GL, et al. Amphiregulin and epiregulin expression in colorectal carcinoma and the correlation with clinicopathological characteristics. Onkologie 2010;33:353-8. [PubMed]
- Kuramochi H, Nakajima G, Kaneko Y, et al. Amphiregulin and Epiregulin mRNA expression in primary colorectal cancer and corresponding liver metastases. BMC Cancer 2012;12:88. [PubMed]
- Prenen H, De Schutter J, Jacobs B, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res 2009;15:3184-8. [PubMed]
- Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 2009;69:1851-7. [PubMed]
- Frattini M, Saletti P, Romagnani E, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer 2007;97:1139-45. [PubMed]
- Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 2009;27:5924-30. [PubMed]
- Jubb AM, Hurwitz HI, Bai W, et al. Impact of vascular endothelial growth factor-A expression, thrombospondin-2 expression, and microvessel density on the treatment effect of bevacizumab in metastatic colorectal cancer. J Clin Oncol 2006;24:217-27. [PubMed]
- Foernzler D, Delmar P, Kockx M, et al. Tumor tissue based biomarker analysis in NO16966: A randomized phase III study of first-line bevacizumab in combination with oxaliplatin-based chemotherapy in patients with mCRC. ASCO Gastrointestinal Cancers Symposium. Orlando, FL, USA, 2010 January 22-24.
- Willett CG, Duda DG, di Tomaso E, et al. Efficacy, safety, and biomarkers of neoadjuvant bevacizumab, radiation therapy, and fluorouracil in rectal cancer: a multidisciplinary phase II study. J Clin Oncol 2009;27:3020-6. [PubMed]
- Jürgensmeier JM, Schmoll HJ, Robertson JD, et al. Prognostic and predictive value of VEGF, sVEGFR-2 and CEA in mCRC studies comparing cediranib, bevacizumab and chemotherapy. Br J Cancer 2013;108:1316-23. [PubMed]
- Kopetz S, Hoff PM, Morris JS, et al. Phase II trial of infusional fluorouracil, irinotecan, and bevacizumab for metastatic colorectal cancer: efficacy and circulating angiogenic biomarkers associated with therapeutic resistance. J Clin Oncol 2010;28:453-9. [PubMed]
- Hegde PS, Jubb AM, Chen D, et al. Predictive impact of circulating vascular endothelial growth factor in four phase III trials evaluating bevacizumab. Clin Cancer Res 2013;19:929-37. [PubMed]
- Gerger A, LaBonte M, Lenz HJ. Molecular predictors of response to antiangiogenesis therapies. Cancer J 2011;17:134-41. [PubMed]
- Estevez-Garcia P, Rivera F, Molina-Pinelo S, et al. Gene expression profile predictive of response to chemotherapy in metastatic colorectal cancer. Oncotarget 2015;6:6151-9. [PubMed]


