
miR-133a-5p suppresses gastric cancer through TCF4 down-regulation
Introduction
Cancer has been the primary cause of death all around the world (1). Gastric cancer (GC), which is the 5th most common cancer and the 3rd leading cause of cancer-related death in the world, seriously threatens the health of people. There are nearly one million people diagnosed with GC every year (2). The prognosis of GC is always poor and the average 5-year survival rate is less than 20% (3), which leads to the urgent importance of exploiting the underlying molecular mechanism and potential drug target for cancer therapy.
MicroRNAs (miRNAs) are a family of endogenous non-coding single-stranded RNA molecules, with a length of 19-22 nucleotides. miRNAs bind to the 3'-untranslated region (UTR) of mRNAs and cause the degradation of targeted mRNAs, leading to the downregulation of protein expression in cytoplasm (4,5). miRNAs are widely found in various biological processes, including tumor progression. miRNAs are highly correlated with the progression of cancer through binding to their targeted genes (4,6,7). For example: miR-21 was an important diagnostic marker for colon and breast cancer, miR-21 contributes to the occurrence of lung cancer; miR-548d serves as a tumor suppressor in of pancreatic cancer; miR-142-5p and miR-375 can be used to predict the recurrent risk for patients with GC (8). Several miRNA-targeting therapeutics have been applied in clinical development, which suggests the elucidation of the underlying mechanism of how miRNAs modulate the process of cancer will contribute to cancer therapy (9).
Recent studies demonstrated that miR-133a-5p was lower expressed and exhibited anti-tumor function in several malignancies, including colorectal cancer, renal cell cancer, bladder cancer, esophageal squamous cell carcinoma, and rhabdomyosarcoma (10-14). Sakr et al. revealed that miR-133a suppressed glioma cell proliferation and migration through targeting membrane-type-1 matrix metalloproteinase (15). Besides, miR-133a inhibited cell proliferation, migration, invasion, and cell cycle progression via targeting transcription factors Sp1 (16). However, there is no study on the effect of miR-133a-5p in the GC profession. In this study, we examined the expression profile and functional roles of miR-133a-5p in GC. Furthermore, we found TCF4 as a novel target gene of miR-133a-5p in GC. Together, miR-133a-5p serves as a tumor suppressor and may play a key role in miRNA-targeted therapeutics for GC.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-20-418).
Methods
Cell culture and transfection
Human gastric epithelial cell line GES-1 and human gastric carcinoma cell line MGC-803, BGC-823, HGC-27, SGC-7901 were all purchased from Xiamen Immocell Biotechnology Co., Ltd (URL: http://immocell.com/html/cn/pc/cn_about.html. Catalog number: IM-H084, IM-H086, IM-H083, IM-H085, IM-H090, respectively). We cultured these cell lines with standard methods in DMEM containing 10% FBS (GIBCO, CA, USA) fetal bovine serum and supplemented with 100 U/mL penicillin and 100 U/mL streptomycin (15140122, GIBCO, CA, USA). Our lab constructed the TCF4 3’UTR WT/Mut plasmid, miR-133a-5p mimics, miR-133a-5p inhibitors, and miRNA NC were synthesized by Ribobio Technology Co., Ltd. (Guangzhou, China).
Bioinformatic analysis
The StarBase (http://starbase.sysu.edu.cn/) database was used to query to miR-133a-5p expression in gastric tumor and tissue. The miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php), TargetScan (http://www.targetscan.org/vert_71/) and miRDB (http://mirdb.org/index.html), was used for the miR-133a-5p predicted target gene and common gene selection. The StarBase database (http://starbase.sysu.edu.cn/) was used for miR-133a-5p and its target genes TCF4 correlation analysis by Spearman tests. The GEPIA2 (http://gepia2.cancer-pku.cn/#index) was used to investigate the TCF4 expression in tumor tissues and normal tissues. The cbioportal (http://www.cbioportal.org/), database analysis of TCF4, and correlation of clinical factors. The Kaplan-Meier plotter prognosis analysis was carried out on the TCF4 (http://kmplot.com/analysis/index) database in GSE14210, GSE15459, GSE22377, GSE29272, GSE51105, GSE62254, six data set of 881 samples of clinical data, draw TCF4 production curve. The GEPIA2 (http://gepia2.cancer-pku.cn/#index) data to the predictive analysis of TCF4, survival curve drawing. The CIBERSORT was used to quantify the cellular immune infiltration and calculate the correlation between TCF4 expression and immune cells. The ESTIMATE package was used to calculate the immune score for each sample and map the correlation between TCF4 and the immune microenvironment. The TCGA pan carcinoma data, extract the expression levels of TCF4 and immune-related genes and draw the heat map of TCF4 and immune-related genes in 33 tumors was downloaded from the UCSC Xena (http://xena.ucsc.edu/) database. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Luciferase-reporter activity assay
For luciferase-reporter activity assay and Western blot analysis, TCF4 3’UTR, WT/Mut, Rab27a 3’UTR, VEGFB 3’UTR, VIM 3’UTR, and miR-133a-5p mimics were transfected into HGC-27 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. miR-133a-5p mimics/inhibitor/NC were transfected into HGC-27 cells using ribo FECT ™ CP Transfection Kit (Ribobio Technology Co., Ltd, Guangzhou, China) according to the manufacturer’s instructions. After a 48-h transfection, cells were used for cell proliferation, EdU incorporation, or trans-well assay.
Quantitative PCR and QuickChange PC
We extracted total RNA from cells using Trizol reagent (Vazyme Biotech Co., Ltd, Nanjing, China) according to the manufacturer’s instructions. For mRNA reverse transcription, cDNA was obtained using a HiScript II One Step qRT-PCR SYBR Green Kit (Q221-01, Vazyme Biotech Co., Ltd.) with 1 ug total mRNA. Moreover, miRNA is reverse transcribed using a miRNA 1st Strand cDNA Synthesis Kit (MR101-01; Vazyme Biotech Co., Ltd.) according to the manufacturer’s instructions. qPCR was performed with primers using ChamQ SYBR Color qPCR Master Mix (Q411-02, Vazyme Biotech Co., Ltd.). Quantification cycle (Cq) values of genes were equilibrated to 18s for mRNA or U6 for miRNA. For wild type and mutation type of TCF4, QuickChange PCR was performed using a PrimeSTAR® HS DNA Polymerase [R010Q, Takara Biomedical Technology (Beijing) Co., Ltd.] according to the manufacturer’s instructions. Each detection was performed in triplicate.
Luciferase-reporter activity assay
The transfected cells were lysed after 48 hours post-transfection, and the luciferase activity was measured using a TransDetect Double-Luciferase Reporter Assay Kit (TransGene Biotech, Beijing, China) and the renilla luciferase activity was used to be the control. Each group was performed in triplicate.
Plasmid construction and lentiviral infection
The lentivirus-based vector pCDH-EF1α-TCF4-BSD plasmid (Antihela) was used for TCF4 overexpression. Lentiviral particles carrying TCF4 were produced as aforementioned. Two days after lentiviral infection, the cells were supported in 4.0 µg/mL blasticidin S (Sigma-Aldrich; Merck KGaA) for ten days to generate stably overexpressing cells.
Western blot analysis
The transfected cells were lysed with a cold RIPA buffer, to measure the protein concentration, we used a BCA Protein Assay Kit (EpiZyme, Shanghai). Equal quantities of proteins (20 μg) are loaded and separated on 10% SDS-PAGE gels and then transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membrane was blocked with 5% BSA (Merck Life Science Co., Ltd., Shanghai, China) at room temperature for 1 hour. Samples were then incubated with primary antibodies against the following proteins: TCF4 (Cat#A1141; 1:2,000; ABclonal, Wuhan, China), and GAPDH (Cat#ab9485; 1:2,000; Abcam, Shanghai, China) at 4 °C overnight. We used horseradish peroxidase-conjugated goat anti-rabbit (Cat#G-21234; 1:10,000; Thermo Fisher Scientific, Waltham, MA, USA) as the secondary polyclonal antibodies, followed by the ECL detection system (Millipore, Billerica, MA, USA) to image protein bands.
Cell proliferation
An MTT assay was performed following the manufacturer’s instructions (Promega, WI, USA). Briefly, the transfected cells were confluent in 96-well cell culture microplates; each of them was seeded in triplicate. And then added 20 μL of MTT solution to each well. The microplate was incubated at 37 °C for 4 hours. After that, 100 μL of DMSO was intercalated to each well. We measured the optical density (OD) at 570 nm using a 96-well plate reader (CMax Plus, Molecular Devices).
EdU incorporation
The transfected cells were incubated at 37 °C for two hours with DMEM containing EdU (50 μm, RiboBio, Guangzhou, China), After that, the cells were fixed with 4% formaldehyde for 20 minutes and then added glycine for 5 minutes. After 10 minutes, it was treated at room temperature with 0.5% Triton X-100. The cells were washed with PBS for three times, and then each well was treated with a 200 μL 1 X Apollo reaction cocktail for 10 minutes. Subsequently, each well was stained with 100 μL of DAPI (5 μg/mL) for 10 minutes and imaged using a fluorescent microscope (MOTIC, Hongkong, China).
Migration and invasion assay
Cell migration and invasion assays were performed using 24-well Transwell chambers (8-μm pores, Thermo Fisher Scientific, Waltham, MA USA). The transfected cells (5×104 cells per well) are cultured with FBS-free medium in the top chamber. And the lower chamber added 600 μL complete media. Twenty-four hours later, we removed the medium from the chamber and the Transwell, and then gently wiped the chamber with a cotton swab. After that, we fixed the migrating cells in 4% paraformaldehyde for 15 min and subsequently stained with crystal violet solution (Cat#ab246820, Abcam) for 5 min. After washing with distilled water to remove excess stain, the migrated cells were photographed under a microscope (MOTIC). The Transwell membranes are precoated with Matrigel (Thermo Fisher Scientific, Waltham, MA USA) to perform the cell invasion assays. The procedure was like cell migration assays. The migrated or invasive cells were counted using software Image J 1.52v (NIH, Bethesda,MD, USA) in three random fields.
Statistical analysis
SPSS software 23.0 (IBM SPSS, Armonk, NY, USA) was used to analyze data. Data are represented as mean ± standard deviation of three independent experiments. Unpaired Student’s t-test was performed to identify significant difference between two groups of data. A value of P<0.05 was statistically significant.
Results
Identification of miR-133a-5p as a tumor suppressor in GC cell lines
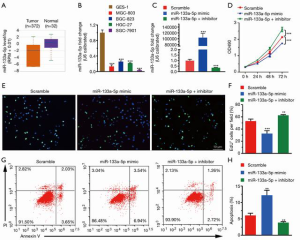
It is well-known that the expression level of genes is strongly associated with their exertive biological functions. Thus, we firstly analyzed the miR-133a-5p level in GC in the TCGA database, where we found that the miR-133a-5p showed reduced expression level in GC (Figure 1A). Also, we used RT-qPCR assay to evaluate the expression levels of miR-133a-5p in different human GC cell lines, including BGC-803, BGC-823, HGC-27, and SGC-7901, and the normal gastric cell line: GES-1. As shown in Figure 1B, the miR-133a-5p expression level was significantly lower than that in the GC cell lines compared to Ges-1. To further confirm the functional roles of miR-133a-5p in GC, the SGC-7901, which displays the lowest expression level of miR-133a-5p, is selected for the following cell function assay model systems in vitro. As shown in Figure 1C, the expression level of miR-133a-5p was up-regulated in miR-133a-5p mimics group and decreased in the transfected group with miR-133a-5p inhibitors, compared to the scramble group. Furthermore, the MTT tests were applied to examine whether the modifications of miR-133a-5p expression can influence cell viability and proliferation. As shown in Figure 1D, the cell viability of SGC-7901 was suppressed in the miR-133a-5p mimics group. However, the miR-133a-5p inhibitors group displayed enhancement of cell viability. These results suggest that miR-133a-5p will decrease cell viability. To further examine whether the miR-133a-5p can inhibit cell proliferation through cell cycle progression, a transient EdU cell proliferation assay was introduced. The EdU cell proliferation assay showed that the population of cells in the S phase was decreased after the transfection of miR-133a-5p mimics and was increased after the transfection of miR-133a-5p inhibitors, which indicated miR-133a-5p could inhibit the proliferation of SGC-7901 cells (Figure 1E,F). To further examine whether the miR-133a-5p can inhibit the cell apoptosis, the AnexinV-PI assay showed that the population of apoptotic cells was increased after the transfection of miR-133a-5p mimics and was decreased after transfection of miR-133a-5p inhibitors (Figure 1G,H).
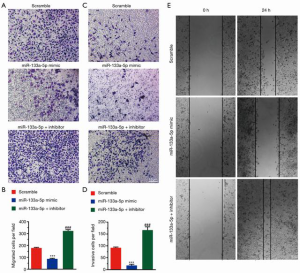
MiR-133a-3p suppresses the migration and invasion of SGC-7901 cells
To investigate the effect of miR-133a-5p on migration and invasion, we performed a Transwell migration and invasion assay. As shown in Figure 2A,B, the number of migrating cells was less in the mimic group than that in the negative control group. Compared with the negative group, the inhibitor group has more migrating cells. Comparable results were found in the Transwell invasion assay (Figure 2C,D). Also, the wound healing assay showed the miR-133a-5p could inhibit the migration of the cells (Figure 2E).
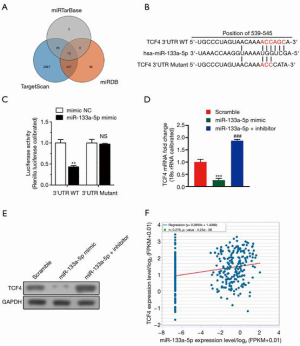
MiR-133a-5p targets TCF4 and down-regulates its expression
Four in silico algorithms (Targetscan, miRanda, and miRwalk) were used to forecast the target genes of miR-133a-5p to elucidate the molecular mechanism by which miR-133a-5p suppresses the tumor growth of GC cells (Figure 3A). The binding sites of the potential candidate: TCF4 was analyzed and found conserved sites (Figure 3B). Next, we applied the dual-luciferase reporter assay system to identify whether they were target genes of miR-133a-5p. Fortunately, we found that the luciferase activity of SGC-7901 cells co-transfected with miR-133a-5p mimics and TCF4-3’-UTR constructs significantly decreased, compared with other control groups (Figure 3C). Furthermore, the luciferase activity was not reduced when the TCF4-3’-UTR construct was a mutant type, which indicated the interaction between TCF4-3’-UTR and miR-133a-5p were necessary for miR-133a-5p’s functions (Figure 3C). Also, qPCR and western blot analysis was used to analyze the effect of miR-133a-5p on the protein level of TCF4. As shown in Figure 3D,E, the mRNA and protein expression level of TCF4 was markedly reduced in SGC-7901 cells transfected with miR-133a-5p mimics but increased after transfection with miR-133a-5p inhibitors (Figure 3D,E). Furthermore, the expression of TCF4 was positively related to the expression of miR-133a-5p (Figure 3F). The results showed that miR-133a-5p directly targeted TCF4 and down-regulated its expression.
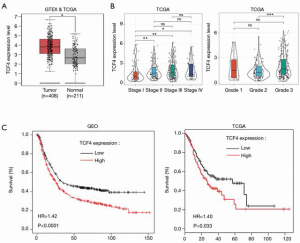
MiR-133a-5p regulated TCF4 as a prognostic gene in GC
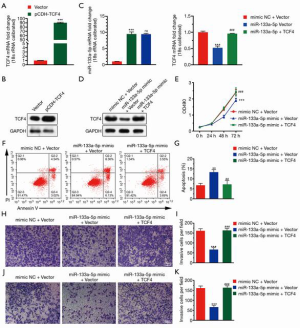
To investigate the functional roles of TCF4 in GC. We used the TCGA database to examine the differential expression of TCF4 in gastric tumor and normal tissue and the expression of TCF4 in different tumor stages (Figure 4A,B). Also, the expression level of TCF4 was positively associated with the survival of the patients in the GEO and TCGA database (Figure 4C). Furthermore, we found that the overexpression of TCF-4 mRNA level in GC cell lines was downregulated (Figure 5A). As shown in Figure 5B, the overexpression of the TCF-4 protein level, was detected in GC cell lines. Interestingly, the mRNA and protein expression level of TCF4 was apposite in cells transfected with miR-133a-5p-mimic and miR-133a-5p-inhibitor, respectively (Figure 5C,D). Besides, TCF4 overexpression rescues the cell proliferation (Figure 5E). Also, the apoptosis of the cells was rescued by TCF4 overexpression (Figure 5F,G). The TCF4 overexpression could also rescue the migration and the invasion effects induced by the miR-133a-5p (Figure 5H,I,J,K).
The potential mechanism by which TCF4 regulates the GC progression
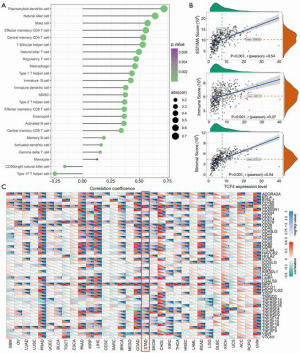
As shown in Figure 6A,B,C, we found that the correlations of the TCF4 mRNA level with the immune cells were significant. Also, the protein-protein interaction, TCF4 mutation genes, TCF4 altered expression analysis was shown in Figure 7.

Discussion
GC is ranked the 3rd leading cause of cancer-related death, with an over 75% mortality rate worldwide (2,17). miRNA is dysregulated in the process of carcinogenesis and is considered useful in the diagnosis and treatment of cancer (8). miR-133a-5p has been identified as a tumor suppressor in several types of cancers such as colorectal cancer, renal cell cancer, bladder cancer, esophageal squamous cell carcinoma, and rhabdomyosarcoma (10-14). In the present study, we found that miR-133a-5p was downregulated in both gastric carcinomas and GC cell lines. Thus, we supposed that miR-133a-5p might also exhibit anti-tumor activity in GC. Therefore, we testify our hypothesis by investigating the cellular function of miR-133a-5p in vitro. Our results revealed that miR-133a-5p targets TCF4 to inhibit cell proliferation and metastasis, and promote cell apoptosis, which indicated that low expression of miR-133a-5p contributed to GC progression. miR-133a-5p may be a potential target for the treatment of gastric cancer.
The TCF/LEF family, a group of transcription factors, bind to DNA using a high mobility group domain. They play essential role in the Wnt signaling pathway, where they recruit the coactivator β-catenin to enhance the transcription of targeting genes, including some oncogene, for example, c-Myc. There are four TCF genes (TCF1, LEF1, TCF3, TCF4) in mammals. TCF4 is important for maintaining the functional property of stem cells (18). Lu et al. have demonstrated that TCF4 promotes proliferation and wound healing in human corneal epithelial stem cells (19). The study of Zhao et al. showed that TCF4 promoted the growth of hepatocellular cancers (20). In our study, we revealed that miR-133a-5p suppresses GC by targeting TCF4. All above suggest that miR-133a-5p may inhibit the progression of GCvia Wnt signaling pathway.
CCND1 is a key protein that regulates the entry of the G0/G1 phase into the S phase and is the well-known targets of TCF4/β-catenin transcription factors complex in the activated Wnt signaling pathway (21). Dong et al. revealed that the transfection of miR-133a improved the population of G0/G1-phase cells (12). Our results of EdU assay showed that the population of S-phase cells decreased after the transfection of miR-133a-5p mimics but increased after the transfection of miR-133a-5p inhibitors. All above suggested that miR-133a-5p downregulates the expression of TCF4 so as to reduce the transcription of CCDN1, which inhibits cell proliferation by inducing arrest at G0/G1 phase.
A miRNA can target numerous genes. Several targets of miR-133a have been reported such as MMP14, Sp1, and SOX4 (16,22,23). High MMP14 expression predicts worse survival in GC (24,25). Sp1 overexpression promotes growth and metastasis of GC (26,27). SOX4 is known as an oncogene in many kinds of tumors including GC (28,29). These indicate that miR-133a-5p exerts its effects through the downregulation of TCF4 as well as by other means.
It may be more meaningful if a ceRNA regulatory network is formed for analysis and research. We used five databases (miRcode, Human LNCediting, miRNet, lncRNASNP2) to find potential lncRNA targeting miR-133a-5p. Lamentedly, there was no related lncRNA. Our next work is to further confirm the conclusion of our study in vivo.
In conclusion, our study confirms the role of miR-133a-5p in GC and the underlying molecular mechanism: miR-133a-5p/TCF4 pathway.
Acknowledgments
Funding: The Wenzhou Science and Technology Project supported this research: Y20160120.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jgo-20-418
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-20-418
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-20-418). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Correa P. Gastric cancer: overview. Gastroenterol Clin North Am 2013;42:211-7. [Crossref] [PubMed]
- Jiang F, Shen XB. miRNA and mRNA expression profiles in gastric cancer patients and the relationship with circRNA. Neoplasma 2019;66:879-86. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172-87. [Crossref] [PubMed]
- Nohata N, Hanazawa T, Kikkawa N, et al. Caveolin-1 mediates tumor cell migration and invasion and its regulation by miR-133a in head and neck squamous cell carcinoma. Int J Oncol 2011;38:209-17. [PubMed]
- Liang H, Hu C, Lin X, et al. MiR-548d-3p Promotes Gastric Cancer by Targeting RSK4. Cancer Manag Res 2020;12:13325-37. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Rao PK, Missiaglia E, Shields L, et al. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J 2010;24:3427-37. [Crossref] [PubMed]
- Kano M, Seki N, Kikkawa N, et al. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer 2010;127:2804-14. [Crossref] [PubMed]
- Dong Y, Zhao J, Wu CW, et al. Tumor suppressor functions of miR-133a in colorectal cancer. Mol Cancer Res 2013;11:1051-60. [Crossref] [PubMed]
- Song T, Xia W, Shao N, et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev 2010;11:905-11. [PubMed]
- Kawakami K, Enokida H, Chiyomaru T, et al. The functional significance of miR-1 and miR-133a in renal cell carcinoma. Eur J Cancer 2012;48:827-36. [Crossref] [PubMed]
- Sakr M, Takino T, Sabit H, et al. miR-150-5p and miR-133a suppress glioma cell proliferation and migration through targeting membrane-type-1 matrix metalloproteinase. Gene 2016;587:155-62. [Crossref] [PubMed]
- Qiu T, Zhou X, Wang J, et al. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett 2014;588:1168-77. [Crossref] [PubMed]
- Wang Z, Liu Q, Huang P, et al. miR-299-3p suppresses cell progression and induces apoptosis by downregulating PAX3 in gastric cancer. Open Life Sci 2021;16:266-76. [Crossref] [PubMed]
- Lu R, Qu Y, Ge J, et al. Transcription factor TCF4 maintains the properties of human corneal epithelial stem cells. Stem cells (Dayton, Ohio) 2012;30:753-61. [Crossref] [PubMed]
- Lu R, Qu Y, Ge J, et al. Transcription factor TCF4 maintains the properties of human corneal epithelial stem cells. Stem Cells 2012;30:753-61. [Crossref] [PubMed]
- Zhao DH, Hong JJ, Guo SY, et al. Aberrant expression and function of TCF4 in the proliferation of hepatocellular carcinoma cell line BEL-7402. Cell Res 2004;14:74-80. [Crossref] [PubMed]
- Zheng L, Liang X, Li S, et al. CHAF1A interacts with TCF4 to promote gastric carcinogenesis via upregulation of c-MYC and CCND1 expression. EBioMedicine 2018;38:69-78. [Crossref] [PubMed]
- Xu M, Wang YZ. miR133a suppresses cell proliferation, migration and invasion in human lung cancer by targeting MMP14. Oncol Rep 2013;30:1398-404. [Crossref] [PubMed]
- Li S, Qin X, Li Y, et al. MiR-133a suppresses the migration and invasion of esophageal cancer cells by targeting the EMT regulator SOX4. Am J Transl Res 2015;7:1390-403. [PubMed]
- Kasurinen A, Gramolelli S, Hagström J, et al. High tissue MMP14 expression predicts worse survival in gastric cancer, particularly with a low PROX1. Cancer Med 2019;8:6995-7005. [Crossref] [PubMed]
- Kasurinen A, Tervahartiala T, Laitinen A, et al. High serum MMP-14 predicts worse survival in gastric cancer. PLoS One 2018;13:e0208800 [Crossref] [PubMed]
- Peng X, Wu M, Liu W, et al. miR-502-5p inhibits the proliferation, migration and invasion of gastric cancer cells by targeting SP1. Oncol Lett 2020;20:2757-62. [Crossref] [PubMed]
- Zhang X, Yang H, Jia Y, et al. circRNA_0005529 facilitates growth and metastasis of gastric cancer via regulating miR-527/Sp1 axis. BMC Mol Cell Biol 2021;22:6. [Crossref] [PubMed]
- Moreno CS. SOX4: The unappreciated oncogene. Semin Cancer Biol 2020;67:57-64. [Crossref] [PubMed]
- Peng X, Liu G, Peng H, et al. SOX4 contributes to TGF-β-induced epithelial-mesenchymal transition and stem cell characteristics of gastric cancer cells. Genes Dis 2018;5:49-61. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)


