
miR-744-5p suppresses tumor proliferation and metastasis by targeting transforming growth factor-beta 1 (TGF-β1) in hepatocellular carcinoma (HCC)
Introduction
Hepatocellular carcinoma (HCC) has been identified as the 6th most common cancer in the world, and has the 4th highest mortality rate among all types of cancer (1). In 2018, an estimated 841,000 new cases of liver cancer were reported globally (1). Currently, there are many treatments for liver cancer, but its prognosis remains unsatisfactory (2,3). HCC remains a great threat and challenge to human health. Thus, new treatments need to be found for HCC urgently.
In a recent study, the development of HCC was found to be closely associated with various genes and abnormal activations of signaling pathways (4). Hepatocarcinogenesis is not only associated with protein-coding genes, but is also involved in non-coding ribonucleic acids (RNAs) (5). MicroRNAs (miRNAs) are endogenous non-coding RNAs that are composed of 18–24 nucleotides (6). It is thought that more than 60% of the genes that encode proteins are managed by miRNAs (5). Recent reports have suggested that miRNAs regulate HCC progression by acting as tumor promoters or suppressors (7-9). For example, miR-140-3p can suppress the expression of epithelial-mesenchymal-transition (EMT)-associated proteins by targeting the mitogen-activated protein kinase (MAPK) signaling pathway (10). MiR-382 has been reported to play an important inhibitory role in non-small cell lung cancer (11). Additionally, miR-494 has been shown to have suppressive effects on epithelial ovarian cancer and may be a new prognostic marker (12). However, the underlying biological function and potential molecular mechanism of miR-744-5p in HCC progression remain an enigma.
Transforming growth factor-beta (TGF-β) is a well-known signaling pathway that is overexpressed in advanced and highly metastatic HCC tissues (13,14). Recent studies have shown that the TGF-β signaling pathway is involved in liver fibrosis and hepatocarcinogenesis (15). Additionally, TGF-β signaling was shown to regulate tumor apoptosis, proliferation, EMT, and invasion in the progression of liver cancer (16-18). Our previous study also showed that TGF-β receptor I serves as a target for miR-4458 and activates drosophila mothers against decapentaplegic2/3 (Smad2/3) and thus mediates HCC development (19).
In this study, we detected miR-744-5p expression in HCC cells and analyzed its effect on the clinical stages of HCC. We found that miR-744-5p suppresses HCC cell proliferation and metastasis. Next, we explored how miR-744-5p regulates HCC progression via the TGF-β1/Smad axis. In addition, the relationships between miR-744-5p and TGF-β1 were validated by a western-blot assay and a luciferase activity reporter assay. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-319).
Methods
Tissue specimens
Tissue specimens were collected from 33 Chinese patients with HCC who underwent hepatectomy for primary HCC at the Department of Hepatobiliary Surgery of the First Affiliated Hospital of Chongqing Medical University between October 2018 and January 2020. This study was approved by the Medical Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (No. 2021-156). All patients gave their informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Cell cultures
The human normal liver cell line L-02 and the HCC cell lines Bel-7402, Huh7, SMMC-7721, LM9, Hep3B, and SK-Hep-1 (SIBS, Shanghai, China) were subsequently cultured in Dulbecco’s minimal essential medium (Gibco) with 10% fetal bovine serum (Gibco, Life Technologies). All cells were incubated in a constant-temperature incubator with 5% carbon dioxide (CO2) at 37 °C.
Transfection
The miRNA mimics of the negative control (NC mimics) and miR-744-5p were obtained from GenePharma (Shanghai, China). The TGF-β1 overexpression plasmid (pcDNA3.1-TGF-β1) and the corresponding empty plasmid (pcDNA3.1-NC) were constructed by HanBio (Shanghai, China). The HCC cells were then transfected with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, USA). The transfected cells were collected after 48 h.
RT-PCR
As per the manufacturer’s instructions, total cellular RNA from the HCC cells was purified using a TRIzol reagent (Invitrogen, California, USA). Single-stranded complementary deoxyribonucleic acids (cDNAs) were reverse-transcribed using a reverse transcription system kit (Takara, Shiga, Japan), and a Hairpin-it miRNA quantitative reverse transcription polymerase chain reaction (qRT-PCR) kit (GenePharma). Next, 2 × SYBR Green qPCR Master Mix (Bimake, Houston, USA) and a Bio-Rad CFX96 real-time PCR detection machine (Bio-Rad Laboratories, Hercules, USA) were used to conduct the qRT-PCR assays. The internal references of miR-744-5p and TGF-β1 were U6 small nuclear RNA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), respectively. The primer sequences were as follows: GAPDH, 5'-CTTTGGTATCGTGGAAGGACTC-3' (F), and 5'-GTAGAGGCAGGGATGATGTTCT-3' (R); miR-744-5p, 5'-AATGCGGGGCTAGGGCTA-3' (F), and 5'-GTGCAGGGTCCGAGGT-3' (R); U6, 5-CAGCACATATACTAAAATTGGAACG-3 (F), and 5-ACGAATTTGCGTGTCATCC-3 (R); TGF-β1: 5'-CGACTCGCCAGAGTGGTTAT-3' (F) and 5'-AGTGAACCCGTTGATGTCCA-3' (R). The assay procedure was as follows: denaturation at 95 °C for 10 min, followed by denaturation at 95 °C for 15 sec over 40 cycles, continuous annealing at 60 °C for 30 s, and elongation at 72 °C for 1 min. The relative quantification expression was calculated using the comparative 2−ΔΔCt method.
Cell-counting kit 8 (CCK-8) assay
To detect cell viability, CCK-8 assays were conducted as directed by the manufacturer (CCK-8 solution; Dojindo, Japan). Transfected HCC cells were seeded into 96-well plates (2×103 cells/well) and cultured in an incubator for 0, 24, 48, and 72 h. The CCK-8 reagent of each well was replaced at different time points and continued to be cultured for 4 h. Finally, we measured the absorbance at 450 nm using the Multiskan Spectrum (Thermo Fisher, Rockford, IL, USA).
Colony-formation assay
Transfected HCC cells were re-seeded onto 6-cm plates with 500 cells/plate and were cultured in a complete medium for 14 days to allow a single cell to grow into a population that could be seen with the naked eye. Next, 3.7% of methanol and 0.1% of crystal violet were used to fix and stain the plate.
Wound-healing assay
The HCC cells were transfected with NC mimics or miR-744-5p mimics and re-seeded into 6-well plates. Next, 10-µL sterile pipette tips were used to make cell scratches. The scratches were captured using a microscope at 200× magnification after phosphate-buffered saline rinsing. Images of the scratches were taken after 24 h of incubation. Finally, the ImageJ system (NIH, Bethesda, USA) was used to analyze the distance that the cell has migrated.
Transwell assay
The ability of the cells to migrate or invade was examined using a Transwell assay. The transfected cells were re-suspended in a serum-free medium and inoculated into the upper part of the 8-µm pore size chambers (Costar, Cambridge, USA), which were either coated or not coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). A 700-µL sample of the complete medium was also added to the bottom of the chamber. After incubation for 24 h, cotton swabs were used to carefully wipe off any non-invaded cells. The cells on the underside of the membrane were then fixed with 95% methanol and stained with crystal violet for 30 min. A Carl Zeiss microscope system (ZEISS, Jena, Germany) was used to randomly capture 3 images. Finally, the number of invaded or migrated cells were counted and averaged across 3 random visual fields.
Dual-luciferase reporter assay
StarBase (http://starbase.sysu.edu.cn/) predicted that TGF-β1 was 1 of the targets of miR-744-5p. A dual-luciferase reporter assay was then conducted to examine the interaction between miR-744-5p and TGF-β1. The mutant (Mut) or wild-type (Wt) TGF-β1 3'-untranslated region (3'-UTR) was inserted into the pSI-Check2 vector (HanBio). Next, miR-744-5p mimics or NC mimics were co-transfected with Wt or Mut TGF-β1 3'-UTR into Hep3B cells. A dual-luciferase reporter system (Promega, Madison, USA) was used to detect luciferase activity after 48 h.
Western-blot assay
After 72 h of transfection, total cellular protein was isolated using sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes. These membranes were incubated in QuickBlockTM blocking buffer (Beyotime, Shanghai, China) for 15 min, then washed with Tris buffered saline with Tween (TBST) solution (20 mM Tris-HCl, 137 mM NaCl, and 0.1% Tween 20). The membranes were then incubated overnight with the following primary antibodies at 4 °C: anti-TGF-β1 (1:1,000, Abways, Shanghai, China), anti-SMAD2 (1:1,000, Abways, Shanghai, China), anti-p-SMAD2 (1:2,000, Abways, Shanghai, China), anti-SMAD3 (1:1,000, Abways, Shanghai, China), anti-p-SMAD3 (1:1,000, Abways, Shanghai, China), anti-E-cadherin (1:2,000, Affinity, Piscataway, USA), anti-N-cadherin (1:1,000, Cell Signaling Technology, Danvers, USA), anti-vimentin (1:1,000, Bimake, Houston, USA), anti-Snail (1:1,000, Cell Signaling Technology, Danvers, USA), and anti-GAPDH (1:20,000, Proteintech, Wuhan, China). Next, the protein bands were washed with TBST buffer and incubated with a secondary antibody. Finally, the membranes were analyzed using a chemiluminescence kit (Merck Millipore, Ltd., Darmstadt, Germany) and Vilber Fusion FX7 (Fusion, FX7, Paris, France).
Statistical analyses
The data are expressed as mean ± standard deviation (SD). The experimental data were statistically analyzed using GraphPad Prism 8.0 software (GraphPad, San Diego, USA). Two groups of normally distributed data were analyzed using the Student’s t-test. A 1-way analysis of variance followed by a Tukey-Kramer test was used to compare 3 or more differences between the treatment groups. Categorical data were tested using a chi-square test. Additionally, the Wilcoxon paired signed-rank test was used to determine the differences between paracancerous tissues and HCC tissues. A P<0.05 was considered statistically significant.
Results
miR-744-5p expression was lower in HCC tissues and cell lines
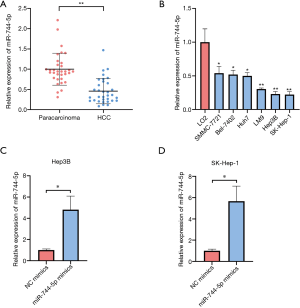
First, we measured the expression levels of miR-744-5p in 33 pairs of clinical samples using the qRT-PCR. As Figure 1A shows, miR-744-5p expression was lower in HCC tissues than adjacent tissues. The pathological characteristics of HCC samples (see Table 1) showed that the expression level of miR-744-5p was significantly associated with liver cirrhosis and microvascular invasion. Additionally, compared to the normal liver cell line L-O2, miR-744-5p expression was significantly lower in HCC cell lines. Notably, miR-744-5p expression was observed to be the lowest in SK-Hep-1 and Hep3B cells (see Figure 1B). Thus, SK-Hep-1 and Hep3B cells were used in this study. Together, these results indicate that miR-744-5p might be relevant to HCC progression.
Full table
miR-744-5p inhibits the proliferation and metastasis of HCC cells
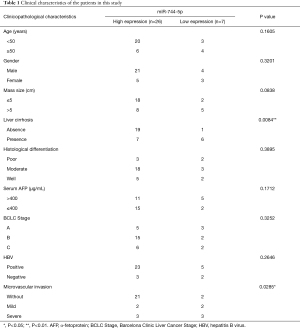
To examine the biological function of miR-744-5p in HCC cells, Hep3B and SK-Hep-1 cells were transfected with miR-744-5p mimics or NC mimics (see Figure 1C,D). The CCK-8 assay results showed that the upregulation of miR-744-5p suppressed the growth of Hep3B and SK-Hep-1 cells (see Figure 2A,B). Additionally, the clone formation assay showed that the miR-744-5p mimics inhibited the proliferation of HCC cells more than the NC mimics (see Figure 2C). The wound-healing assay showed that cell motility significantly decreased after transfection with miR-744-5p mimics (see Figure 2D,E). The Transwell results showed that an overexpression of miR-744-5p decreased the number of Hep3B and SK-Hep-1 cells passing through the Transwell chamber (see Figure 2F,G). As EMT is a well-known factor associated with tumor metastasis, we assessed the main protein of EMT by means of western blotting, and found that miR-744-5p enhanced epithelial markers but reduced the mesenchymal markers (see Figure 2H).
miR-744-5p regulates the development of HCC by targeting TGF-β1 directly
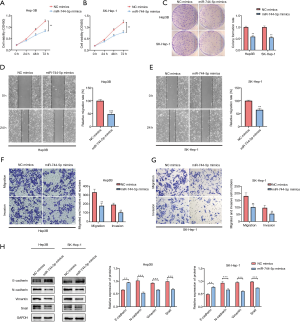
To further explore the potential molecular mechanisms by which miR-744-5p suppresses HCC progression, online public databases were searched to determine the downstream effects of miR-744-5p, and TGF-β1 was identified as the possible target of miR-744-5p (see Figure 3A). The relationship between miR-744-5p and TGF-β1 was examined using a dual-luciferase reporter gene assay. As Figure 3B shows, the luciferase activity was obviously inhibited following co-transfection with miR-744-5p mimics and h-TGF-β1-wt. Conversely, the luciferase activity remained unchanged when the cells were co-transfected with miR-744-5p mimics and h-TGF-β1-mut. Notably, the qRT-PCR results indicated that miR-744-5p might cause an obvious decrease in TGF-β1 expression in Hep3B and SK-Hep-1 cells, which also suggested that TGF-β1 might act as a possible target of miR-744-5p (see Figure 3C,D). Subsequently, western blotting was used to analyze the effects of miR-744-5p on the TGF-β1 signaling pathway. The expressions of TGF-β1, p-Smad2, and p-Smad3 were observed to decrease significantly after miR-744-5p mimics were transfected into Hep3B and SK-Hep-1 cells (see Figure 3E,F).
TGF-β1 overexpression abrogates the tumor suppressive effect of miR-744-5p in HCC development
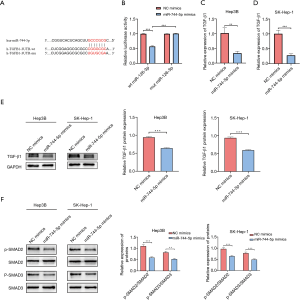
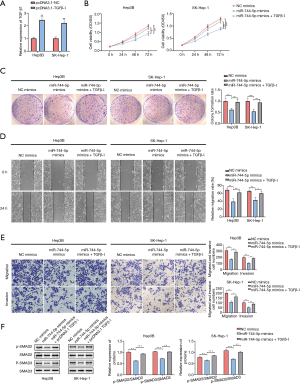
To determine whether miR-744-5p, which regulates the biological function of HCC cells, was mediated by TGF-β1, pcDNA3.1 was used to upregulate the expression of TGF-β1 in Hep3B and SK-Hep-1 cells (see Figure 4A). As expected, the CCK-8 and the clone-formation assays showed that overexpressed TGF-β1 could partially restore the suppression effect of miR-744-5p mimics in HCC cell growth (see Figure 4B,C). Additionally, wound-healing (see Figure 4D) and Transwell assays (see Figure 4E) showed that TGF-β1 overexpression could abrogate the inhibitory effect of miR-744-5p mimics on invasion and migration. The results of the western blotting showed that the overexpression of both miR-744-5p and TGF-β1 led to the recovery of p-SAMD2 and p-SMAD3 protein levels (see Figure 4F).
Discussion
In recent decades, great advances have been made in the treatment of liver cancer. However, the prognosis of patients with liver cancer continues to be poor and unsatisfactory, which can be attributed to its high metastasis rate and frequent recurrence (20). Thus, exploring the details of HCC development and the pathogenesis of HCC is an effective means of improving the quality of life of patients. Recently, miRNAs have been the focus of research due to their critical role in tumor progression (21,22). Our results showed that miR-744-5p was more lowly expressed in HCC than para-carcinoma tissues. Additionally, the low expression of miR-744-5p was found to be closely correlated with microvascular invasion. miR-744-5p has also been found to suppress HCC cell proliferation and metastasis. Thus, we concluded that miR-744-5p plays a vital role in the development of liver cancer.
It is currently believed that high differentiation, low mortality, and a strong invasion and metastasis ability are the significant characteristics of tumor cells. These processes are often accompanied by the abnormal activation of many signaling pathways (23). Thus, finding the downstream target of miR-744-5p is essential if the occurrence and development of HCC is to be elucidated. In recent reports, miR-744-5p was identified as a diagnostic marker and therapeutic target for cancers due to its ability to target ADP ribosylation factor 1 (ARF1) (24), paired box 2 (PAX2) (25), heterogeneous nuclear ribonucleoprotein C (HNRNPC) (26), etc. However, in our study, it was predicted through starBase that TGF-β1 may be the downstream target of miR-744-5p. The luciferase reporter showed that miR-744-5p could target TGF-β1 directly.
Over the years, TGF-β1 signaling has been studied extensively; however, the role of TGF-β1 signaling in HCC remains controversial. TGF-β1 signaling has been shown to play a tumor suppressor role in the early phases of tumors development (27). However, TGF-β1 signaling promotes EMT and induces tumor cell invasion in the later phases (28). TGF-β1 signaling is also reported to promote angiogenesis during HCC development (29). In this study, we showed that the overexpression of TGF-β1 largely prevented the inhibition effects of the miR-744-5p mimics on the biological functions of HCC cells. Further, the miR-744-5p mimics were found to regulate SMAD2/3 expression, which has been considered as the downstream target of TGF-β1. These results indicated that the anti-cancer effects of miR-744-5p could be increased by targeting the TGF-β1 signaling pathway.
In conclusion, our study showed that the expression of miR-744-5p was highly repressed in HCC tissues and cell lines. Further functional experiments showed that miR-744-5p exerts a tumor-suppressive role in HCC by directly targeting TGF-β1. However, the effects of the miR-744-5p/TGF-β1 pathway on HCC growth in vivo need to be further explored in future studies. Together, these findings provide new insights into the mechanisms involved in the occurrence and development of HCC. Further, these findings may have identified a prognostic marker that can be used as an important therapeutic target in the treatment of liver cancer.
Acknowledgments
Funding: This work was supported by the Venture and Innovation Support Program for Chongqing Overseas Returnees [CX2017105, CX2018146], and the Postgraduate Innovation Project of Chongqing Education Commission, Chongqing, China [CYS19184].
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-319
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-319
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-319). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Medical Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (No. 2021-156). All patients gave their informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Bruix J, Han KH, Gores G, et al. Liver cancer: Approaching a personalized care. J Hepatol 2015;62:S144-56. [Crossref] [PubMed]
- El-Serag HB, Marrero JA, Rudolph L, et al. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology 2008;134:1752-63. [Crossref] [PubMed]
- Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, et al. Hepatocellular carcinoma: From diagnosis to treatment. Surg Oncol 2016;25:74-85. [Crossref] [PubMed]
- Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med 2018;16:220. [Crossref] [PubMed]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 2019;20:5-20. [Crossref] [PubMed]
- Yuan Q, Loya K, Rani B, et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology 2013;57:299-310. [Crossref] [PubMed]
- Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res 2016;76:3666-70. [Crossref] [PubMed]
- Markopoulos GS, Roupakia E, Tokamani M, et al. A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol (Dordr) 2017;40:303-39. [Crossref] [PubMed]
- Zhang QY, Men CJ, Ding XW. Upregulation of microRNA-140-3p inhibits epithelial-mesenchymal transition, invasion, and metastasis of hepatocellular carcinoma through inactivation of the MAPK signaling pathway by targeting GRN. J Cell Biochem 2019;120:14885-98. [Crossref] [PubMed]
- Chen D, Zhang Y, Lin Y, et al. MicroRNA-382 inhibits cancer cell growth and metastasis in NSCLC via targeting LMO3. Exp Ther Med 2019;17:2417-24. [Crossref] [PubMed]
- Yuan J, Wang K, Xi M. MiR-494 Inhibits Epithelial Ovarian Cancer Growth by Targeting c-Myc. Med Sci Monit 2016;22:617-24. [Crossref] [PubMed]
- Lin TH, Shao YY, Chan SY, et al. High Serum Transforming Growth Factor-β1 Levels Predict Outcome in Hepatocellular Carcinoma Patients Treated with Sorafenib. Clin Cancer Res 2015;21:3678-84. [Crossref] [PubMed]
- Watanabe Y, Iwamura A, Shimada YJ, et al. Transforming Growth Factor-β1 as a Predictor for the Development of Hepatocellular Carcinoma: A Nested Case-Controlled Study. EBioMedicine 2016;12:68-71. [Crossref] [PubMed]
- Fabregat I, Caballero-Díaz D. Transforming Growth Factor-β-Induced Cell Plasticity in Liver Fibrosis and Hepatocarcinogenesis. Front Oncol 2018;8:357. [Crossref] [PubMed]
- Dooley S, ten Dijke P. TGF-β in progression of liver disease. Cell Tissue Res 2012;347:245-56. [Crossref] [PubMed]
- Giannelli G, Mikulits W, Dooley S, et al. The rationale for targeting TGF-β in chronic liver diseases. Eur J Clin Invest 2016;46:349-61. [Crossref] [PubMed]
- Dituri F, Mancarella S, Cigliano A, et al. TGF-β as Multifaceted Orchestrator in HCC Progression: Signaling, EMT, Immune Microenvironment, and Novel Therapeutic Perspectives. Semin Liver Dis 2019;39:53-69. [Crossref] [PubMed]
- Zhang Y, Shi K, Liu H, et al. miR-4458 inhibits the epithelial-mesenchymal transition of hepatocellular carcinoma cells by suppressing the TGF-β signaling pathway via targeting TGFBR1. Acta Biochim Biophys Sin (Shanghai) 2020;52:554-62. [Crossref] [PubMed]
- Zhang G, Li R, Deng Y, et al. Conditional survival of patients with hepatocellular carcinoma: results from the Surveillance, Epidemiology, and End Results registry. Expert Rev Gastroenterol Hepatol 2018;12:515-23. [Crossref] [PubMed]
- Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 2018;15:137-51. [Crossref] [PubMed]
- Anwar SL, Lehmann U. MicroRNAs: Emerging Novel Clinical Biomarkers for Hepatocellular Carcinomas. J Clin Med 2015;4:1631-50. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Zhao LG, Wang J, Li J, et al. miR-744-5p inhibits cellular proliferation and invasion via targeting ARF1 in epithelial ovarian cancer. Kaohsiung J Med Sci 2020;36:799-807. [Crossref] [PubMed]
- Chen S, Shi F, Zhang W, et al. miR-744-5p Inhibits Non-Small Cell Lung Cancer Proliferation and Invasion by Directly Targeting PAX2. Technol Cancer Res Treat 2019;18:1533033819876913 [Crossref] [PubMed]
- Kleemann M, Schneider H, Unger K, et al. MiR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells. Sci Rep 2018;8:9020. [Crossref] [PubMed]
- Hu G, Niu F, Humburg BA, et al. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget 2018;9:18648-63. [Crossref] [PubMed]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131-42. [Crossref] [PubMed]
- Mazzocca A, Fransvea E, Lavezzari G, et al. Inhibition of transforming growth factor beta receptor I kinase blocks hepatocellular carcinoma growth through neo-angiogenesis regulation. Hepatology 2009;50:1140-51. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


