
Increased RBM12 expression predicts poor prognosis in hepatocellular carcinoma based on bioinformatics
Introduction
In recent decades, liver cancer has become one of the most common cancers in the world and has a high patient fatality rate. In comparison to developed countries, China has both a higher incidence and higher fatality rate of cancer. Surgery is the most common treatment for hepatocellular carcinoma (HCC), and fluorouracil, sorafenib, and other chemotherapeutic medications being commonly administered (1). Despite a decrease in the mortality rate of patients diagnosed in the early stages of the disease’s progression, the overall survival (OS) for advanced-stage liver cancer patients can still be improved (2). Patients are frequently identified with advanced-stage liver cancer and are unable to obtain appropriate therapy because of a lack of highly specific and specific early diagnostic biomarkers, and thus their prognosis is poor (3). It is critical to investigate new HCC markers to enhance the prognosis of HCC patients, as well as to better understand the molecular pathways driving carcinogenesis and cancer progression.
RNA-binding proteins (RBPs) are proteins that bind RNA via 1 or more globular RNA-binding domains (RBDs) or motifs (RBMs) and can alter the function of the RNAs they bind (4). Various studies have revealed that RBPs are important regulators of posttranscriptional gene expression, and influence RNA modification and stability, including the processes of RNA splicing, localization, translation, and turnover (5). To date, more than 800 messenger RNA (mRNA) RBPs have been identified, but only a small percentage have been characterized in terms of function (6).
RBP dysregulation has been linked to the development of liver tumors and the progression of cancer (7-11). For example, RNA-binding motif protein 43 (RBM43) is downregulated in cases of malignancy where low expression is linked to a poor prognosis for HCC patients (7) Furthermore, in vitro experiments showed that RBM38 can induce apoptosis and senescence in liver cancer cells by inhibiting proliferation and colony development and suppressing migration and invasion (10). RNA-binding motif protein 12 (RBM12) is an RBP belonging to the RBM family. It is a conserved protein that is widely produced in cells. Five different RBMs, two proline-rich regions, and many potential transmembrane domains are found in the RBM12 protein (12). RBM12 was first discovered to have an abnormal expression in meibomian cell carcinoma (13), and its disruption has been linked to psychosis and endometrial cancer (14,15). Despite these discoveries, the specific role of RBM12 in HCC is still unknown.
To determine the clinical significance of RBM12 in HCC, our study used online databases to examine the expression of RBM12 and its predictive value in patients. We also looked at the expression of RBM12 in the cells or tissues of liver cancer, and the functional network of RBM12 was analyzed using LinkedOmics and gene set enrichment analysis (GSEA). Next, we examined the impact of DNA methylation on RBM12, and finally, the link between RBM12 and immune infiltration in HCC was elucidated. Consequently, our study is the first to explore the expression, prognostic significance, immune cell infiltration, and biological networks of RBM12, as well as the probable causes of the overexpression of RBM12 in HCC. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-390).
Methods
Tumor IMmune Estimation Resource (TIMER)
The TIMER (https://cistrome.shinyapps.io/timer) is a tool used for analyzing the infiltration of tumors by immune cells (16). This resource can be used to assess six types of immune cell infiltrates (CD4+ T cells, B cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells). The TIMER was also used to reveal the RBM12 expression in tumors compared with its expression in normal tissues and the distribution of gene expression in mutant and wild-type tumors in HCC.
The Human Protein Atlas
Immunohistochemistry (IHC) data from 44 distinct normal tissue types were located in the Human Protein Atlas (https://www.proteinatlas.org) (17). During this search, we were able to identify the expression data of RBM12 in several human tissues.
UALCAN database analysis
The clinical data of 33 different cancer patients were located on the UALCAN database (http://ualcan.path.uab.edu) (18), which was further used to investigate the relationship between RBM12 and other clinical features in HCC patients.
Kaplan-Meier plotter database analysis
The Kaplan-Meier plotter database (http://kmplot.com/analysis/) includes 5,143 samples of breast cancer, 2,437 samples of lung cancer, 1,816 samples of ovarian cancer, and 1,065 samples of stomach cancer (19). In providing various cancer samples, this database is useful for examining the relationship of genes with patient survival. Our study used the Kaplan-Meier plotter to assess the association between RBM12 and the survival of patients with HCC.
LinkedOmics
LinkedOmics (http://linkedomics.org/login.php) is a free online database that contains information on the 32 cancer types that are also found in The Cancer Genome Atlas (TCGA) (20). LinkFinder, LinkInterpreter, and LinkCompare are the three analytical modules on the website. We used the LinkInterpreter module for an enrichment analysis that was based on Gene Ontology (GO), network modules, and biological pathways in order to gain new biological insights about RBM12 in HCC.
GSEA
GSEA is a computer tool used to determine if there are statistical and consistent differences in a group of genes among two biological states. GSEA was used to analyze TCGA gene expression data from 374 liver cancer samples acquired from the UCSC Xena Functional Genomics Explorer so that the biological processes enriched by RBM12 could be identified.
Databases related to DNA methylation
MEXPRESS (https://mexpress.be/about.html) is a data visualization tool that can present TCGA expressions, clinical data, and DNA methylation, as well as their interconnections (21). TCGA, the International Cancer Genome Consortium (ICGC), and the Gene Expression Omnibus (GEO) have all contributed cancer genomic databases to the cBioPortal (https://www.cbioportal.org/) (22). The link between RBM12 mRNA expression and DNA methyltransferase (DNMT) expression was found via Gene Expression Profiling Interactive Analysis 2 (GEPIA2) (http://gepia.cancer-pku.cn/index.html) (23), while the Shiny Methylation Analysis Resource Tool (SMART) app (http://www.bioinfo-zs.com/smartapp/) is a user-friendly web application used to evaluate TCGA DNA methylation data in detail (24).
Immune infiltrates analysis
We employed CIBERSORT, a deconvolutional technique based on gene expression (http://cibersort.stanford.edu/), to assess the gene expression variations among sample sets (25). We assessed the immune response of 22 tumor-infiltrating immune cells (TIICs), using CIBERSORT to clarify the link between RBM12 expression in HCC and any correlations among the TIICs.
Quantitative real-time polymerase chain reaction (qRT-PCR) and western blot analyses
The TRIzol reagent (Invitrogen Life Technologies, USA) was used for extraction of total RNA from both HCC cells (Huh7, LM3, MHCC97H) and the normal cell line of the liver, known as QSG-7701. To produce complementary DNA (cDNA), a RevertAid First Strand cDNA Synthesis Kit (TaKaRa Bio, Japan) was used for RNA reverse transcription. The primer sequences we used for qRT-PCR analysis are presented in Table 1. qRT-PCR analysis was completed on a LightCycler 480 II (Roche) PCR platform using a SYBR Master Mixture (TaKaRa Bio) and SYBR Master Mixture (TaKaRa Bio) according to the manufacturer’s instructions. To assess the transcriptional expression, the fold change in the expression of RBM12 compared to β-actin was employed (RBM12 expression levels were determined using the −∆Ct method), while RBM12 protein expression in normal and HCC cell lines was determined by western blot analysis. The primary antibody was antihuman RBM12 (1:1,000, sc-514258, Santa Cruz Biotechnology, USA), the secondary antibody was anti-mouse immunoglobulin G (IgG; 1:2,000; A0216, Beyotime, China), and the loading control was β-actin (1:1,000; AF0003, Beyotime, China).

Full table
Cell culture and patients
HCC tissues and corresponding nearby noncancerous tissues were obtained from 45 patients (33 men and 12 women) who had not undergone therapy before surgery at the Affiliated Hospital of Nantong University between 2018 and 2020. The included patients gave their informed consent, and the Affiliated Hospital of Nantong University’s Ethics Committee approved the study (No. 2018-L006). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The samples were promptly frozen in liquid nitrogen and preserved at −80 °C for subsequent use. The Shanghai Institute of Cell Biology (Shanghai, China) provided the HCC cell lines (Huh7, LM3, and MHCC97H), and Beyotime provided a normal liver cell line (QSG-7701; Shanghai, China). Cells were grown in RPMI 1640 medium or Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific), penicillin (100 U/mL), and streptomycin (100 µg/mL) at 37 °C in a 5% CO2 environment.
Statistical analysis
All statistical analyses were conducted using SPSS 22.0 (IBM Corporation, USA) and GraphPad Prism 8.02. The mean and the standard deviation (SD) of the data were calculated, and differences between the sets of data were assessed using a one-way analysis of variance (ANOVA). All experiments were carried out in triplicate.
Results
Expression levels of RBM12 in different types of tissues and tumors
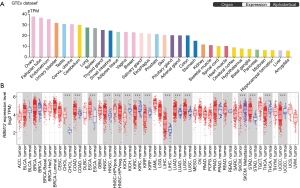
RBM12 expression levels in normal human tissues were discovered by searching The Human Protein Atlas database. The results showed a lower level of RBM12 in the normal liver tissues (Figure 1A). The TIMER was also employed to visualize RBM12 expression in distinct malignancies. Figure 1B describes the high RBM12 expression in liver HCC (LIHC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and stomach adenocarcinoma (STAD). Finally, we discovered that, with the exception of the amygdala, the expression of RBM12 was normally lowest in the liver but elevated in liver cancer.
RBM12 expression in HCC cells and patients
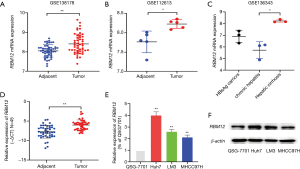
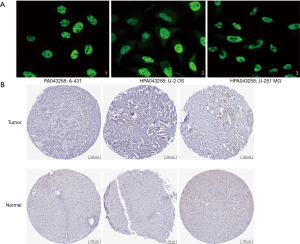
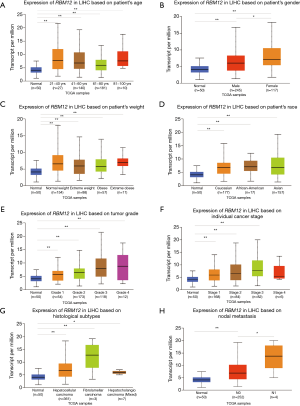
The GSE138178 and GSE112613 data sets indicated higher expressions of RBM12 in HCC (Figure 2A,2B). In addition, we found a significantly higher expression of RBM12 in the liver tissues of patients with cirrhosis than in patients with chronic hepatitis, but the expression showed no significant difference when compared to hepatitis B surface antigen (HBsAg) carriers (Figure 2C). During surgery, 45 paired samples were collected from liver cancer and nearby noncancerous tissues to corroborate these findings. The expression of RBM12 mRNA in these samples was then analyzed using qRT-PCR, revealing a substantial increase of RBM12 expression in tissues of liver cancer as compared to adjacent healthy tissues (Figure 2D). We also looked at RBM12 mRNA levels in the Huh7, LM3, and MHCC97H HCC cell lines and in QSG-7701 (the normal cell line of the human liver; Figure 2E,2F). RBM12 is significantly increased in HCC cells. Indirect immunofluorescence showed RBM12 localized to the nucleoplasm (Figure 3A). IHC staining revealed high expression levels of RBM12 in tumor tissues (Figure 3B), and RBM12 levels were considerably greater in certain categories, such as age, gender, weight, race, tumor grade and stage, histological subtypes, and nodal metastatic status (Figure 4). Cumulatively, these findings show that RBM12 is typically increased during HCC development.
Prognostic potential of RBM12 in HCC
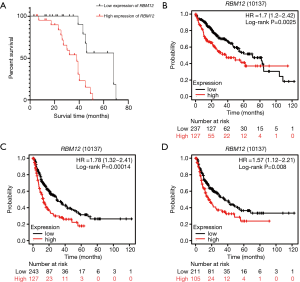
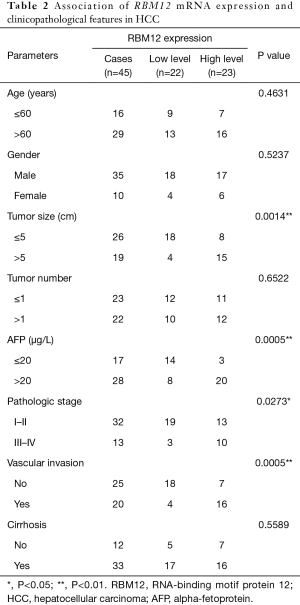
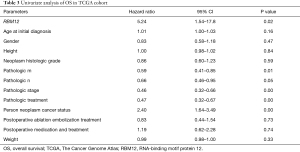
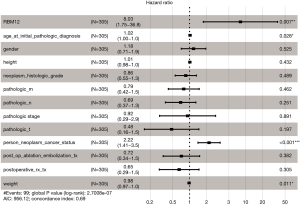
Our results suggested that enhanced RBM12 expression is linked with poor patient prognosis (Figure 5A). Using the Kaplan-Meier plotter database, we examined the connection of RBM12 expression with clinical features, which revealed that RBM12 expression was linked to OS, progression-free survival (PFS), and relapse-free survival (RFS) in HCC patients (Figure 5B-5D). The clinical data of the 45 patients with liver cancer showed that RBM12 was related to tumor size, alpha-fetoprotein (AFP) tumor marker status, pathologic stage, and vascular invasion (Table 2). The possibility of RBM12 serving as a predictive biomarker was confirmed through univariate analysis. As shown in Table 3, RBM12 had prognostic significance in TCGA (P=0.02) cohorts and was related to pathologic stage (P=0.00) and neoplasm cancer status (P=0.00). The multivariate analysis indicated that RBM12 expression was related to age (P=0.028), neoplasm cancer status (P<0.001), and weight (P=0.011; Figure 6). These finding suggest that RBM12 may have impact on the prognosis of patients with liver cancer.
Full table
Full table
LinkedOmics predicts functional annotation and signaling pathways
LinkedOmics was also used to find coexpressed RBM12 genes in the LIHC cohort. Figure 7A displays the significant genes, with a heat map depicting the top 50 most significant genes that have a negative or positive connection with RBM12 expression (Figure 7B,7C). Figure 7D depicts the RBM12 network and 100 coexpressed genes, and the findings of functional enrichment studies of these 100 implicated genes are displayed in Figure 7E. Significant genes were inextricably involved in the cell cycle, peptidyl-lysine modification, BRCA1-BARD1-BACH1-DNA damage complex II, and nuclear export.
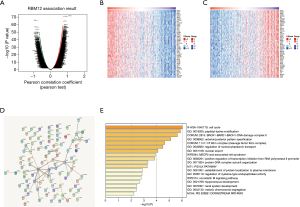
Significant genes and pathways identified by GSEA
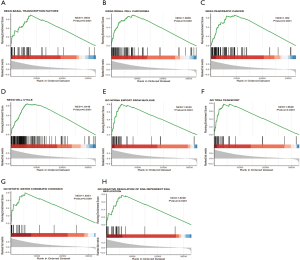
GSEA is one of the more commonly used analysis methods to study the biological function of tumors, as it can detect expression variations in gene sets rather than individual genes, and it is more reliable and flexible than are the more traditional methods like GO and Kyoto Encyclopedia of Genes and Genomes (KEGG). GSEA retrieved the expression data of RBM12 mRNA for HCC samples from TCGA data sets. This data allowed us to better understand the biological relevance of coregulated proteins, with GSEA revealing that RBM12 is involved in a variety of tumor progression pathways, including basal transcription factors, renal cell carcinoma, cell cycle, pancreatic cancer, and noncoding RNA (ncRNA) export from the nucleus (Figure 8). The major bile acid biosynthesis and the flavonoid metabolic pathway were found to be among the gene sets enriched in the low expression phenotypic group (Table 4).

Full table
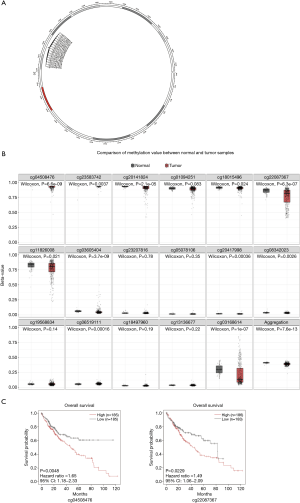
DNA methylation affects RBM12 mRNA levels
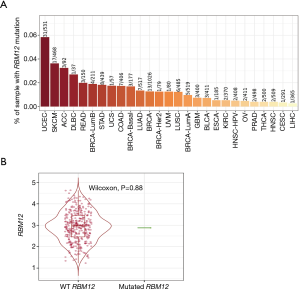
RBM12 gene amplification is rare in HCC (Figure 9A,9B), which implies that other processes are responsible for the enhanced RBM12 expression in HCC. Promoter hypomethylation contributes to tumorigenesis via the transcriptional activation of oncogenes.
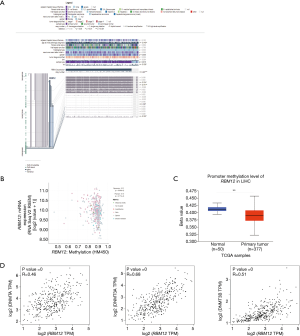
MEXPRESS was used to investigate the methylation status of the RBM12 gene in HCC. Many methylation sites were discovered in the promoter region of the RBM12 gene (Figure 10A). Using the cBioPortal online tool, we discovered a negative relationship between RBM12 mRNA and promoter methylation in HCC (Figure 10B). According to an HCC cohort analysis from TCGA, RBM12 promoter methylation in HCC tissues was considerably lower when compared to normal pancreas tissues (Figure 10C).We further discovered a link between RBM12 mRNA expression and DNMT mRNA expression levels in HCC patients (Figure 10D). Figure 11A visualizes the chromosomal distribution of every 5'-C-phosphate-G-3' (CpG) associated with the RBM12 gene in HCC, and Figure 11B,C show the differences in CpG expression between normal tissue and tumor tissue samples and its effect on prognosis. Our findings suggested that RBM12 is upregulated in HCC due to abnormal promoter methylation.
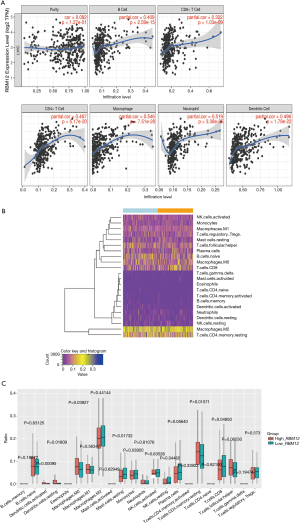
The link between RBM12 and immune cell infiltration in HCC
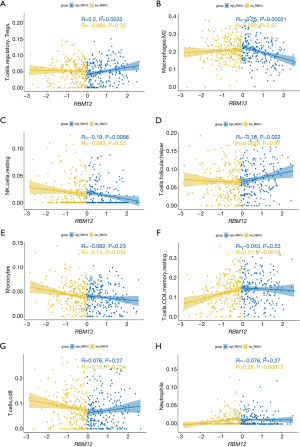
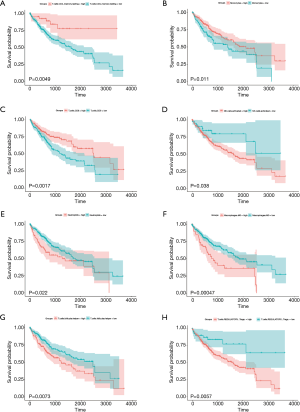
The TIMER database analysis indicated the expression of RBM12 to be substantially correlated with different types of immune cell infiltration (Figure 12A). We aimed to determine whether the tumor immune microenvironment in liver cancer with a high level of RBM12 was different from that with a low level. Based on RBM12, the 374 tumor samples were divided into two groups, with 187 samples in the group of high expression and 187 samples in the group of low expression. In order to determine the levels of the 22 different types of immune cells, we used CIBERSORT to analyze the gene expression of the downloaded samples. The CIBERSORT algorithm, which was applied to the 22 immune cell subtypes, assisted in determining differences in the expression levels of the cell subtypes between the high and low RBM12 expression groups. It was found that the ratios of resting mast cells, monocytes, and CD8+ T cells were significantly higher in the low expression group. Conversely, the proportions of M0 macrophages and resting memory CD4+ T cells were significantly higher in the high expression group (Figure 12B,12C). Furthermore, we demonstrated the correlation between several immune cells and RBM12 in HCC (Figure 13). In 22 types of immune cells, the levels of highly activated natural killer (NK) cells, neutrophils, M0 macrophages, follicular helper T cells, and regulatory T cells level were related to poor OS, while increased resting memory CD4+ T cells, monocytes, and CD8+ T cells were related to better OS (Figure 14).
Discussion
Mortality rates from liver cancer are high all around the world (26). Despite the accessibility of several successful clinical treatments, patient OS rates remain low (2). The majority of people with early-stage liver cancer exhibit no typical symptoms. AFP positivity is found in fewer than 70% of liver cancer patients, and is frequently employed as a diagnostic tool for the early detection of HCC (27). Nonalcoholic fatty liver disease (NAFLD) has become more prevalent due to modern lifestyles and dietary habits. Furthermore, effective hepatitis virus vaccination treatment procedures have become more accepted, and the etiology of liver illnesses has changed, the metabolic risk factors of HCC, including metabolic syndrome, obesity, type II diabetes and NAFLD, are increasing and may become the main causes of HCC (26). In light of these characteristics, novel HCC markers are required to improve the early detection rates of the disease.
Despite the ubiquity of HCC, the pathogenic mechanisms that cause it are still unknown (28). RBPs, which regulate gene expression at the posttranscriptional level (4), are important in the development of HCC (7-11), while the core protein, RBM12 has been linked to the onset of a variety of diseases (13-15). Even though these contributing factors have been recognized by the scientific community, no in-depth research into the role of RBM12 in HCC has been conducted. Thus, a bioinformatics RBM12 analysis of liver cancer was performed in this study.
In database study, RBM12 was found to be overexpressed at the mRNA level in cirrhosis and HCC tissues. In addition, qRT-PCR, western blotting, and sample immunochemistry analysis suggested the upregulation of RBM12 in HCC. Furthermore, RBM12 was shown to be positively linked to age, gender, weight, race, grade, stage, status for nodal metastasis, and histological subtypes. RBM12 was also shown to have a high diagnostic value in the case of HCC, with high levels of RBM12 being connected to poorer OS, PFS, and RFS. Furthermore, univariate and multivariate analyses of clinical data obtained from TCGA revealed significant differences in RBM12 expression during the pathologic stage (P<0.00), in neoplasm cancer status (P<0.00), age (P=0.028) and weight (P=0.011). As a result, we believe RBM12 is a useful diagnostic and prognostic marker for liver cancer.
A functional module from LinkedOmics was used to investigate the RBM12 coexpression mode in the LIHC cohort. According to GO terms, RBM12 coexpressed genes primarily participate in the cell cycle, peptidyl-lysine modification, BRCA1-BARD1-BACH1-DNA damage complex II, and nuclear export. We also analyzed TCGA data on liver cancer patients to discover the expression changes in RBM12 gene sets. The results demonstrated that RBM12 is involved in various pathways, including basal transcription factors, the cell cycle, and ncRNA export from the nucleus. Our findings suggest that elevated levels of RBM12 in HCC patients may affect both cell cycle control and nuclear export. In multiple networks of genes coexpressed with RBM12, cell cycle regulation was a primary enrichment pathway. Cell cycle disruptions induce unlimited proliferation, which is an underlying mechanism of HCC (29,30). Whether RBM12 affects a patient’s prognosis by regulating the cell cycle remains to be further studied. The export of mRNA from the nucleus to the cytoplasm is an important regulatory phase in protein expression. Abnormal RNA export has been observed in primary human cancer specimens, and these cargo RNAs code for proteins that are involved in almost every aspect of cancer (31). RBM12 is most likely involved in these processes, but more research is needed to determine the exact mechanisms of its involvement.
According to TCGA data, RBM12 gene amplification is uncommon in HCC, implying that alternative processes are responsible for the elevated RBM12 expression in HCC (32). Promoter hypomethylation contributes to tumorigenesis through the transcriptional activation of oncogenes (33), and in HCC, DNA methylation changes play a crucial role in regulating pathological and physiological processes (34). We investigated the DNA methylation status of RBM12 in HCC and discovered that RBM12 expression was closely correlated with DNMT expression. Moreover, several methylated CpG sites were significantly correlated with the outcomes of HCC patients. Epigenetic markers are established dynamically and reversibly, and epigenome-targeted medicines are also expanding the therapeutic portfolio for solid tumors (35). Patients with various forms of cancer, including HCC patients, have been treated solely with DNMT inhibitors or a combination of DNMT inhibitors and immunotherapy (36). Our findings suggest that DNA changes that affect the transcriptional levels of the RBM12 gene may influence HCC carcinogenesis, implying that DNMT inhibitors could be an effective therapy for HCC patients.
Tumor immunotherapy is a relatively recent treatment option that is gaining credibility in the context of HCC treatment (37,38). Immunotherapy is a promising area of research into the tumor microenvironment for clinicians in search of therapeutic targets and biomarkers for prognosing and diagnosing HCC (39). The TIMER database was used in our research to find links between immune infiltration levels and RBM12 expression in liver cancer, and according to our findings, the associations of RBM12 with dendritic cells, macrophages, B cells, and T cells are the strongest. Furthermore, our CIBERSORT analysis demonstrated a link between RBM12 expression and immune cell infiltration. Our results show that the levels of M0 macrophages and resting memory CD4+ T cells were significantly higher in the high expression group, whereas the levels of resting mast cells, monocytes, and CD8+ T cells were significantly higher in the low expression group. In HCC patients, immune cell infiltration has a significant impact on survival (40), and we showed that increased abundance of resting memory CD4+ T cell, monocytes, and CD8+ T cells may be related to better OS. Taken together, these findings show that RBM12 is crucial to HCC immune infiltration, cell regulation, and recruitment.
Conclusions
Overall, this is the first study to establish RBM12 as a novel HCC biomarker. Our findings revealed that RBM12 levels are considerably higher in HCC, with high RBM12 levels being predictive of poor prognosis. LinkedOmics and GSEA analysis found RBM12 to be significantly associated with tumor progression, particularly in regard to basal transcription factors, the cell cycle, and nuclear export. In addition, we found that abnormal DNA methylation may increase oncogenic RBM12 expression. In conclusion, our findings show that RBM12 is a suitable target for liver cancer immunotherapy.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (No. 81871927). Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_2798).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-390
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-390
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-390). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to this study are appropriately investigated and resolved. The included patients gave their informed consent, and the Ethics Committee of the Affiliated Hospital of Nantong University approved the study (No. 2018-L006). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. Adv Cancer Res 2021;149:1-61. [Crossref] [PubMed]
- Li S, Saviano A, Erstad DJ, et al. Risk factors, pathogenesis, and strategies for hepatocellular carcinoma prevention: emphasis on secondary prevention and its translational challenges. J Clin Med 2020;9:3817. [Crossref] [PubMed]
- Peng JL, Wu JZ, Li GJ, et al. Identification of potential biomarkers of peripheral blood mononuclear cell in hepatocellular carcinoma using bioinformatic analysis: a protocol for systematic review and meta-analysis. Medicine (Baltimore) 2021;100:e24172 [Crossref] [PubMed]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet 2014;15:829-45. [Crossref] [PubMed]
- Hentze MW, Castello A, Schwarzl T, et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 2018;19:327-41. [Crossref] [PubMed]
- Castello A, Fischer B, Eichelbaum K, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012;149:1393-406. [Crossref] [PubMed]
- Feng H, Liu J, Qiu Y, et al. RNA-binding motif protein 43 (RBM43) suppresses hepatocellular carcinoma progression through modulation of cyclin B1 expression. Oncogene 2020;39:5495-506. [Crossref] [PubMed]
- Lin Y, Liang R, Qiu Y, et al. Expression and gene regulation network of RBM8A in hepatocellular carcinoma based on data mining. Aging (Albany NY) 2019;11:423-47. [Crossref] [PubMed]
- Miao X, Zhang N. Role of RBM3 in the regulation of cell proliferation in hepatocellular carcinoma. Exp Mol Pathol 2020;117:104546 [Crossref] [PubMed]
- Ye J, Liang R, Bai T, et al. RBM38 plays a tumor-suppressor role via stabilizing the p53-mdm2 loop function in hepatocellular carcinoma. J Exp Clin Cancer Res 2018;37:212. [Crossref] [PubMed]
- Chua HH, Tsuei DJ, Lee PH, et al. RBMY, a novel inhibitor of glycogen synthase kinase 3β, increases tumor stemness and predicts poor prognosis of hepatocellular carcinoma. Hepatology 2015;62:1480-96. [Crossref] [PubMed]
- Yang W, Ng P, Zhao M, et al. Promoter-sharing by different genes in human genome--CPNE1 and RBM12 gene pair as an example. BMC Genomics 2008;9:456. [Crossref] [PubMed]
- Kumar A, Kumar Dorairaj S, Prabhakaran VC, et al. Identification of genes associated with tumorigenesis of meibomian cell carcinoma by microarray analysis. Genomics 2007;90:559-66. [Crossref] [PubMed]
- Steinberg S, Gudmundsdottir S, Sveinbjornsson G, et al. Truncating mutations in RBM12 are associated with psychosis. Nat Genet 2017;49:1251-4. [Crossref] [PubMed]
- Shivakumar M, Miller JE, Dasari VR, et al. Exome-wide rare variant analysis from the DiscovEHR study identifies novel candidate predisposition genes for endometrial cancer. Front Oncol 2019;9:574. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14 [Crossref] [PubMed]
- Thul PJ, Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci 2018;27:233-44. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci 2018;5:181006 [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Koch A, De Meyer T, Jeschke J, et al. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics 2015;16:636. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102 [Crossref] [PubMed]
- Li Y, Ge D, Lu C. The SMART App: an interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin 2019;12:71. [Crossref] [PubMed]
- Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773-82. [Crossref] [PubMed]
- McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021;73:4-13. [Crossref] [PubMed]
- Luo P, Yin P, Hua R, et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology 2018;67:662-75. [Crossref] [PubMed]
- Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. [Crossref] [PubMed]
- Chen Z, Xie H, Hu M, et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res 2020;10:2993-3036. [PubMed]
- Li Q, Sun M, Wang M, et al. Dysregulation of Wnt/β-catenin signaling by protein kinases in hepatocellular carcinoma and its therapeutic application. Cancer Sci 2021;112:1695-706. [Crossref] [PubMed]
- Borden KLB. The nuclear pore complex and mRNA export in cancer. Cancers (Basel) 2020;13:42. [Crossref] [PubMed]
- Sato N, Maitra A, Fukushima N, et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res 2003;63:4158-66. [PubMed]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27-56. [Crossref] [PubMed]
- Wang M, Ye Q, Mao D, et al. Research progress in liver-regenerating microenvironment and DNA methylation in hepatocellular carcinoma: the role of traditional Chinese medicine. Med Sci Monit 2020;26:e920310 [Crossref] [PubMed]
- Thiagalingam S. Epigenetic memory in development and disease: Unraveling the mechanism. Biochim Biophys Acta Rev Cancer 2020;1873:188349 [Crossref] [PubMed]
- Huang X, Yang C, Wang J, et al. Integrative analysis of DNA methylation and gene expression reveals distinct hepatocellular carcinoma subtypes with therapeutic implications. Aging (Albany NY) 2020;12:4970-95. [Crossref] [PubMed]
- Hilmi M, Vienot A, Rousseau B, et al. Immune therapy for liver cancers. Cancers (Basel) 2019;12:77. [Crossref] [PubMed]
- Nishida N, Kudo M. Immune phenotype and immune checkpoint inhibitors for the treatment of human hepatocellular carcinoma. Cancers (Basel) 2020;12:1274. [Crossref] [PubMed]
- Federico P, Petrillo A, Giordano P, et al. Immune checkpoint inhibitors in hepatocellular carcinoma: current status and novel perspectives. Cancers (Basel) 2020;12:3025. [Crossref] [PubMed]
- Schoenberg MB, Li X, Li X, et al. The predictive value of tumor infiltrating leukocytes in hepatocellular carcinoma: a systematic review and meta-analysis. Eur J Surg Oncol 2021; Epub ahead of print. [Crossref] [PubMed]
(English Language Editors: J. Collie and J. Gray)


