
FAT10 promotes hepatocellular carcinoma (HCC) carcinogenesis by mediating P53 degradation and acts as a prognostic indicator of HCC
Introduction
Hepatocellular carcinoma (HCC), a major type of liver cancer, is one of the ten most common cancers globally. It ranks as the fifth leading cause of cancer death in males, and it accounts for 70% to 85% of the total cancer burden (1-3). Despite the increasingly diverse and individualized treatment of HCC, it is difficult to obtain a substantial improvement in the five-year survival rate of patients. This is due to the high heterogeneity of HCC, which is already at an advanced stage of the disease at the time of diagnosis, and it seriously threatens people’s lives and health (4,5). Therefore, it is urgent to elucidate the important molecules responsible for hepatocarcinogenesis and determine the prognosis of HCC so that novel strategies for HCC prevention and treatment can be developed (6). Up to now, a variety of oncogenes and abnormally activated cell survival or proliferation signals including c-Met, nuclear factor-κB (NF-κB), Wnt/β-catenin, and c-Jun N-terminal kinase (JNK) pathways have been detected in human liver cancer samples. At the same time, the overexpression of oncogenes and the over-regulation of proliferation signals eventually lead to cell canceration (7-10). However, some studies have shown that the downregulation of anti-oncogenic molecules in hepatocytes, including p53 and HNF-4α, also promotes liver carcinogenesis or increases cell proliferation (11,12). These results reveal the complexity of HCC pathogenesis. Herein, this study reports a novel HCC oncogene, FAT10, and its role in hepatocarcinogenesis. Furthermore, it documents a mechanism which FAT10 interacts with and mediates the degradation of p53, which acts as an important tumor suppressor gene. And the relationship between the expression of FAT10 and the prognosis of HCC patients. This study is expected to discover new combined targets and prognostic warning markers for HCC treatment. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-374).
Methods
Ethical statement
The clinicopathological characteristics of all patients are shown in Table 1. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics boards of the Seventh People’s Hospital of Shanghai University of Traditional Chinese Medicine, the First Affiliated Hospital of China Medical University and the Third Affiliated Hospital of Shanghai Military Medical University (NO.: 2020-7th-HIRB-018, [2021]2021-223-2 and EHBHKY2020-K-040) and informed consent was taken from all the patients. The animal studies involved in this experiment were performed after approval by the ethics committee of the General Hospital of Northern Theater Command. All animal experiments were performed at the laboratory animal center of the General Hospital of Northern Theater Command in accordance with international guidelines for biomedical research involving animals. A protocol was prepared before the study without registration.
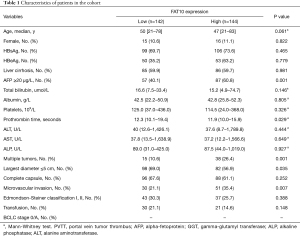
Full table
Histological examination and tissue microarray analysis
A tissue microarray block containing 4 kinds of digestive system tumor tissues was purchased from Outdo Biotech Co., Ltd. (Shanghai, China). HCC tissues were obtained from surgical resection specimens of HCC patients from the Seventh People’s Hospital of Shanghai University of Traditional Chinese Medicine, the First Affiliated Hospital of China Medical University and Eastern Hepatobiliary Surgery Hospital. Immunohistochemistry was performed following standard protocols. The antibodies used are listed in Table S1. The assessment of tissue microarray staining was based on the percentage of positively stained cells, and the staining intensity was measured using ImageScope software (Aperio Technologies, Inc., Vista, CA, USA).
Experimental animal models
Diethylnitrosamine (DEN) was administered to 30 male Wistar rats through intraperitoneal injections once a week for 10 weeks. To monitor the development of HCC, 2 rats were sacrificed every 2 weeks after DEN administration. The remaining rats were killed 22 weeks after the first DEN administration. The 6-week-old male C57 mice used in the experiment were purchased from Shanghai SLAC Laboratory Animal Co., Ltd., Chinese Academy of Sciences. They were reared in accordance with the specific pathogen-free (SPF)-level laboratory animal breeding standards, with an ambient temperature of approximately 25 °C and relative humidity of approximately 60%. They were reared at 5 mice per cage, with a circadian rhythm of 12/12 hours and free food and water intake. After the purchase, the experiment was carried out after breeding in the animal room for more than 1 week. Construction of carbon tetrachloride (CCl4) and bile duct ligation (BDL)—induced murine liver cirrhosis model was performed as described in previous papers (13-15). After the model was successfully established, it was followed by histological evaluation.
Cell lines
The HCC cell line SMMC-7721 was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences.
Cell proliferation assay
SMMC-7721-FAT10 and control cells were seeded in 96-well plates and incubated for different periods. Then, Cell Counting Kit-8 reagent (Dojindo, Kumamoto, Japan) was added to the cells, and adenosine triphosphate (ATP) activity was measured on a Synergy 2 microplate reader (BioTek, Winooski, VT, USA).
Real-time polymerase chain reaction (PCR)
An ABI PRISM 7300 Sequence Detector (Applied Biosystems, Foster City, CA, USA) was used to perform real-time fluorescent quantitative PCR reactions and detect the expression level of each molecule.
The sequence of each primer is shown in Table S2.
Pathway analysis of FAT10 mediated HCC invasion and metastasis
The gene expression profifiles of HCC were downloaded from TCGA database, which was contained 424 samples, including 50 normal samples and 374 tumor samples. R language (Limma package and Beeswarm package) was used to conduct single gene difference analysis of FAT10, Gene Set Enrichment Analysis was performed using the GSEA software (gsea-3.0-jar) (https://www.broadinstitute.org/gsea/) with gene set database = c2 kegg, permutation = phenotype.
Statistical analysis
Overall survival (OS) is defined herein as the time from the day of surgery to death (including any cause). Qualitative variables were compared using the c2 test or Fisher’s exact test, and continuous variables were compared using the student t-test. Correlation analysis between FAT10 expression and sample survival time was performed by the Kaplan Meier method and compared with the log rank test. Cox proportional hazards models were used to determine independent factors for survival and recurrence. All statistical analyses were performed using SPSS version 18.0 (SPSS Inc., SPSS Statistics for Windows, Chicago, IL, USA). P<0.05 was considered statistically significant in this study.
Results
FAT10 expression is elevated in human HCC
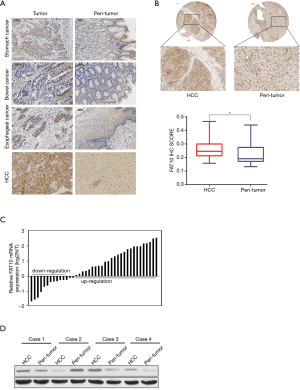
To evaluate the expression of FAT10 among various digestive system tumors, FAT10 expression was first evaluated in digestive cancer samples. Based on an array containing 4 digestive cancer samples from patients and corresponding noncancerous tissues for immunohistochemical staining, it was found that FAT10 expression was up-regulated in HCC (Figure 1A). Next, the elevated expression of FAT10 in human liver cancer tissues was confirmed (246/286) (Figure 1B). It was further observed that FAT10 transcripts in HCC (105/134) were significantly increased compared to adjacent tissues (Figure 1C) and western blotting results (Figure 1D) also showed that FAT10 was up-regulated.
FAT10 overexpression mediates poor prognosis of HCC patients
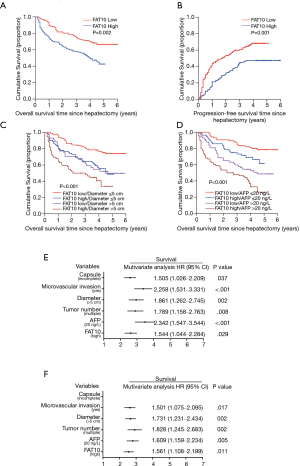
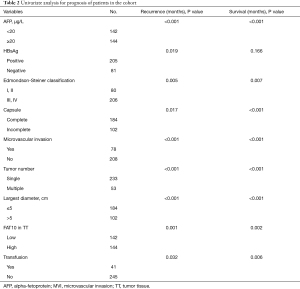
The test results of a cohort of 286 HCC patients indicated an increase in FAT10 expression. According to the results of immunostaining, 286 HCC patients were divided into the FAT10 high expression (FAT10 high) group (n=144) and FAT10 low expression (FAT10 low) group (n=142). Compared with the low FAT10 group, the OS of HCC patients in the high FAT10 group was also shorter; the median OS time for the FAT10 low and FAT10 high group was 46 and 15 months, respectively, with a difference of 31 months (P=0.002). Compared with the low FAT10 group, the Progression-free survival (PFS) of HCC patients in the high FAT10 group was shorter.; the median PFS times for the for the FAT10 low and FAT10 high group were 26 and 15 months, respectively, with a difference of 11 months (P=0.001) (Figure 2A,B, Table 1). A clinical cohort study showed that FAT10 overexpression in tumors with a diameter >5 cm and alpha-fetoprotein (AFP) >20 ng/L displayed an even worse prognosis in terms of OS time (Figure 2C,D). In multivariate analysis, it was proved that capsule, vascular invasion, diameter, tumor number, AFP, and FAT10 overexpression are independent prognostic factors that affect OS time and recurrence time of patients (Figure 2E,F, Tables 2,3). Thus, the overexpression of FAT10 may be an important early warning factor that mediates the low survival rate of HCC patients and the poor prognosis.
Full table
Full table
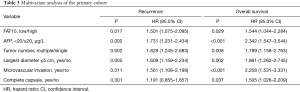
Expression of FAT10 is elevated during hepatocarcinogenesis
In order to explore the expression of FAT10 in the process of HCC, liver samples were collected from Wistar rats where DEN had induced hepatocarcinogenesis, and then the samples were used for immunohistochemical staining. The rats gradually showed fibrosis and cirrhosis in the liver after receiving DEN administration, and ultimately all rats progressed to HCC in the 22nd week after treatment. Intriguingly, the expression of FAT10 was significantly increased during the process of liver cancer in rats (Figure 3A-C). Next, a CCl4 and BDL-triggered mouse cirrhosis model was established, and then FAT10 expression was assessed (Figure 3D-G). The final results showed that, compared with normal tissues, the expression of FAT10 in the fibrotic or cirrhotic liver was significantly increased.
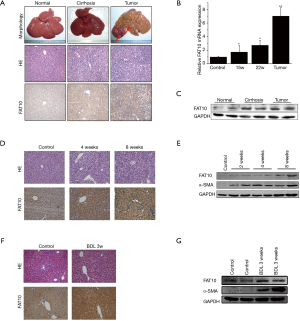
FAT10 mediates p53 degradation and promotes hepatoma cell proliferation
This study used pc3-FAT10 (FAT10 expression vector) or scrambled control plasmid to transform SMMC-7721 cells to clarify further the role of FAT10 in the malignant biological behavior of hepatoma cells. As shown in Figure 4A and 4B, FAT10 overexpression remarkably enhanced the proliferation of HCC cells and led to the formation of more colonies. To determine the molecular mechanism underlying the FAT10-induced cell proliferation and hepatocarcinogenesis, the expression of key tumor-related genes was determined. Notably, no significant difference was detected in the expression of the β-catenin or HIF-1 reporter genes in FAT10 overexpression cells (Figure 4C,D). Nevertheless, the expression of the p53 reporter gene was found to be decreased by FAT10 overexpression (Figure 4E). Subsequently, FAT10 and p53 expression were analyzed in HCC patients, and the results revealed a possible negative correlation between FAT10 and p53 (Figure 4F,G). Furthermore, SMMC-7721 cells were treated with cycloheximide (CHX), and it was found that CHX did not affect reducing p53 expression mediated by FAT10. However, the application of MG132 could significantly block this down-regulation, indicating that FAT10 promotes p53 degradation-dependent mechanisms through the proteasome (Figure 4H,I).
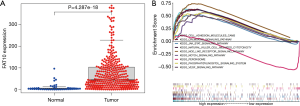
FAT10 is involved in the regulation of invasion and metastasis pathways of liver cancer
The expression of FAT10 in HCC was higher than that in normal tissues, and the difference was significant (P<0.05) (Figure 5A). A total of 84 FAT10 related pathways were obtained by GSEA pathway enrichment analysis (P<0.05). There were 9 pathways related to metastasis of liver cancer including: nod like receptor signaling pathway, chemokine signaling pathway, cell adhesion molecules cams and so on (Figure 5B).
Discussion
HCC has a high incidence and is the most common fatal malignant tumor globally (16). People have been seeking to improve screening, diagnosis and treatment strategies to improve the prognosis of this malignant tumor. Effective biomarkers for monitoring the prognosis of HCC are still relatively lacking. At present, studies have shown that AFP, alpha-fetoprotein LP-3 (AFP-L3), DCP, HGF and other molecules are closely related to the poor prognosis of HCC vascular invasion and distant metastasis (17-20). In recent years, studies on the relationship between non-coding RNA, immune-related molecules, and key factors in serum exosomes and the prognosis of HCC have gradually increased (21-23). Therefore, screening reliable molecular markers that can evaluate the prognosis of HCC patients and evaluate the efficacy will have important clinical translational value. So far, surgical resection is recognized as the most effective therapeutic method. However, according to clinical study results, surgical resection only prolongs the lifespan of early HCC patients (24). How to choose better treatments to prevent recurrence has become a challenge that affects the OS of HCC patients. The formation of HCC patients with medication plays a key role in preventing recurrence. In view of this, comprehensive, sequential, and individualized comprehensive treatment can bring more clinical benefits to patients (25). At present, targeted therapy is still the first choice for HCC treatment. It is a treatment plan for point-to-point killing, inactivation, and inhibition of the promoter genes of liver cancer to make it lose its function (24,26). And the wide application of immunotherapy has brought new hope for HCC patients. The unique immunobiology of the liver promotes tumorigenesis and tumor immune tolerance under conditions of fibrosis and chronic inflammation, and also provides the possibility of immune targeted therapy. In addition to tumor cells, the tumor microenvironment of HCC is composed of stromal cells (including immune cells, fibroblasts, endothelial cells, etc.), structural components (such as extracellular matrix, etc.), and signal components (chemokines, growth factors, etc.) (27). These components affect tumor immune escape, response to immunotherapy and patient prognosis. Part of the dysfunction of effector cells in the HCC microenvironment is directly driven by HCC cells, and the other part is indirectly driven by suppressive immune cells recruited into the tumor (28). Compared with other tumors, the tumor microenvironment of HCC also has a certain degree of particularity. The liver is a special immune organ that contains a large number of antigen presenting cells (APCs) and immune cells. Due to long-term exposure to chronic antigens from the intestine, the liver has a certain degree of immune tolerance (29). In addition, most HCCs in my country are accompanied by chronic hepatitis B virus (HBV) infection. HBV infection interacts with other components of the tumor microenvironment in complex signaling pathways, resulting in the special tumor microenvironment of HCC (30).
Given the previous positive results of critical oncogene identification for cancer therapy, this study examined the role of FAT10 and its tumor-promoting effects (31,32). Substantial genetic and biochemical data support the notion that FAT10 has a role in promoting tumors. FAT10, also called ubiquitin D, belongs to the ubiquitin protein family. It is covalently bound to target proteins, leading to their degradation by the proteasome (33,34). Previous studies have reported its role in the immune system, in which it promotes the degradation of I kappa B alpha. FAT10 is involved in the lipopolysaccharide (LPS)-induced activation of TNF alpha or the intrinsic immune response, and FAT10 may participate in the maturation of dendritic cells (35,36). Furthermore, FAT10 was found to have a close relationship with metastasis and tumor, nodes, and metastases (TNM) staging in human gastric cancer (37). It has been shown that FAT10 plays an important role in HCC development by a variety of regulatory mechanisms. Liu et al. (38) illuminated that effects of FAT10 is may be its influence on the Akt/GSK3b pathway to promote the progression of hepatitis B-related HCC, increasing the levels of cyclin D1 and c-myc protein. Yan et al. (39) found that FAT10 promotes HCC proliferation via the WISP1 protein degradation pathway. Gao et al. (40) indicated that FAT10 activates NF-κB, which in turn upregulated the chemokine receptors CXCR4 and CXCR7 result in the HCC cell malignant biological behavior. Based on the existing studies, pro-inflammatory cytokine interferon (IFN-γ) and tumor necrosis factor (TNF-α) can jointly upregulate FAT10 expression in HCC cells by activating the p65/NF-κB pathway and increasing the expression of IFN-γ-and TNF-α-dependent proteasal subunit LMP2 (36,41). While TNF-α and interleukin (IL-6) increase FAT10 expression by the transcription factors STAT3 and the p65/NF-κB pathway (42,43). Glucose-regulated protein 78 (GRP78) and ubiquitin-like protein FAT10 can each promote the occurrence of HCC (44,45). However, studies have shown that GRP78 can increase the expression of FAT10 through the NF-κB pathway, thereby promoting the proliferation of HCC (46). This study clarified the new mechanism of GRP78 to promote the progress of HCC, analyzed the relationship between GRP78 and FAT10 in HCC, provided a new idea for the study of the mechanism of HCC, and provided a new target for the prevention and treatment of HCC.
In this study, through in-depth exploration of HCC samples, it was confirmed that FAT10 increased significantly at the tissue level, gene level, and protein level. This increase in expression can mediate worse survival prognosis and malignant clinicopathological characteristics of HCC patients. Furthermore, using multivariate analysis, it was found that FAT10 is an effective independent risk factor that affects the OS rate hazard ratio (HR) value after curative resection. More importantly, FAT10 overexpression enhanced the accuracy of predicting OS outcomes in patients with HCC. Moreover, it was observed that FAT10 expression increased in the gradual progression of carcinogenesis in the DEN-induced rat HCC and BDL or CCl4 induced liver cirrhosis models, which means that FAT10 is involved in the occurrence of hepatocarcinogenesis. Accordingly, this study clarified that FAT10 plays a critical role in HCC carcinogenesis in animal models and human patients. To gain insights into the mechanisms regulating HCC carcinogenesis, HCC cell lines were transformed with plasmids to overexpress FAT10. It was further found that FAT10 promoted HCC cell proliferation and clone formation, which implied a proto-oncogenic function in promoting tumor carcinogenesis. Interestingly, this study determined that FAT10 acts on P53. The down-regulation of p53 mediated by FAT10 can regulate cell proliferation and clone formation, which plays an important role in the carcinogenesis of HCC (47).
The above fully demonstrates that FAT10 can mediate the down-regulation of p53 expression, thereby providing new insights into the clinical significance of individualized therapy and prognosis prediction for HCC patients. In summary, it is reasonable to believe that FAT10 functions as a proto-oncogene and acts as a critical prognostic indicator of HCC. However, the specific signal pathway needs to be further studied.
Acknowledgments
The authors would like to thank Yun-en Liu for her technical assistance and sincerely thank Jin Ding and HY Wang for their selfless guidance. In addition, the mouse model and rat liver fibrosis, and the paraffin samples and pictures of liver cirrhosis models are presented by Dr. Ding Jin and Dr. Sun Wen from the Eastern Hepatobiliary Surgery Hospital.
Funding: This work was supported by grants from the Key Projects of Liaoning Natural Science Foundation (20180530019); Natural Fund Guidance Program of Liaoning Province (2019-ZD-1072); Major scientific and technological innovation research and development plan (19-112-4-079); China Medical University Youth Backbone Support Program (1210519002); Shanxi Administration of Traditional Chinese Medicine (2019-GJ-JC005).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-374
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-374
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-374). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics boards of the Seventh People’s Hospital of Shanghai University of Traditional Chinese Medicine, the First Affiliated Hospital of China Medical University and the Third Affiliated Hospital of Shanghai Military Medical University (NO.: 2020-7th-HIRB-018, [2021]2021-223-2 and EHBHKY2020-K-040) and informed consent was taken from all the patients. The animal studies involved in this experiment were performed after approval by the ethics committee of the General Hospital of Northern Theater Command. All animal experiments were performed at the laboratory animal center of the General Hospital of Northern Theater Command in accordance with international guidelines for biomedical research involving animals. A protocol was prepared before the study without registration.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Fan M, Qian N, Dai G. Expression and prognostic significance of doublecortin-like kinase 1 in patients with hepatocellular carcinoma. Oncol Lett 2017;14:7529-37. [Crossref] [PubMed]
- Huber AR, Gonzalez RS, Orloff MS, et al. Accuracy of vascular invasion reporting in hepatocellular carcinoma before and after implementation of subspecialty surgical pathology sign-out. Indian J Pathol Microbiol 2017;60:501-4. [Crossref] [PubMed]
- Kim NH, Lee T, Cho YK, et al. Impact of clinically evident portal hypertension on clinical outcome of patients with hepatocellular carcinoma treated by transarterial chemoembolization. J Gastroenterol Hepatol 2018;33:1397-406. [Crossref] [PubMed]
- Mazzaferro V, Sposito C, Zhou J, et al. Metroticket 2.0 Model for Analysis of Competing Risks of Death After Liver Transplantation for Hepatocellular Carcinoma. Gastroenterology 2018;154:128-39. [Crossref] [PubMed]
- Niu L, Liu L, Yang S, et al. New insights into sorafenib resistance in hepatocellular carcinoma: Responsible mechanisms and promising strategies. Biochim Biophys Acta Rev Cancer 2017;1868:564-70. [Crossref] [PubMed]
- Bouattour M, Raymond E, Qin S, et al. Recent developments of c-Met as a therapeutic target in hepatocellular carcinoma. Hepatology 2018;67:1132-49. [Crossref] [PubMed]
- Kim E, Lisby A, Ma C, et al. Promotion of growth factor signaling as a critical function of β-catenin during HCC progression. Nat Commun 2019;10:1909. [Crossref] [PubMed]
- Ng PKS, Lau CPY, Lam EKY, et al. Hypermethylation of NF-κB-Activating Protein-Like (NKAPL) Promoter in Hepatocellular Carcinoma Suppresses Its Expression and Predicts a Poor Prognosis. Dig Dis Sci 2018;63:676-86. [Crossref] [PubMed]
- Lin S, Hoffmann K, Gao C, et al. Melatonin promotes sorafenib-induced apoptosis through synergistic activation of JNK/c-jun pathway in human hepatocellular carcinoma. J Pineal Res 2017; [Crossref] [PubMed]
- Li Z, Han C, Feng J. Relationship of the expression levels of XIAP and p53 genes in hepatocellular carcinoma and the prognosis of patients. Oncol Lett 2017;14:4037-42. [Crossref] [PubMed]
- Morimoto A, Kannari M, Tsuchida Y, et al. An HNF4α-microRNA-194/192 signaling axis maintains hepatic cell function. J Biol Chem 2017;292:10574-85. [Crossref] [PubMed]
- Liu Q, Shen WF, Sun HY, et al. Hydrogen-rich saline protects against liver injury in rats with obstructive jaundice. Liver Int 2010;30:958-68. [Crossref] [PubMed]
- Sun H, Chen L, Zhou W, et al. The protective role of hydrogen-rich saline in experimental liver injury in mice. J Hepatol 2011;54:471-80. [Crossref] [PubMed]
- Xu WH, Hu HG, Tian Y, et al. Bioactive compound reveals a novel function for ribosomal protein S5 in hepatic stellate cell activation and hepatic fibrosis. Hepatology 2014;60:648-60. [Crossref] [PubMed]
- Han T, Xiang DM, Sun W, et al. PTPN11/Shp2 overexpression enhances liver cancer progression and predicts poor prognosis of patients. J Hepatol 2015;63:651-60. [Crossref] [PubMed]
- Bruix J, Cheng AL, Meinhardt G, et al. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: Analysis of two phase III studies. J Hepatol 2017;67:999-1008. [Crossref] [PubMed]
- Saito Y, Shimada M, Utsunomiya T, et al. Prediction of recurrence of hepatocellular carcinoma after curative hepatectomy using preoperative Lens culinaris agglutinin-reactive fraction of alpha-fetoprotein. Hepatol Res 2012;42:887-94. [Crossref] [PubMed]
- Poté N, Cauchy F, Albuquerque M, et al. Performance of PIVKA-II for early hepatocellular carcinoma diagnosis and prediction of microvascular invasion. J Hepatol 2015;62:848-54. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Guo CM, Liu SQ, Sun MZ. miR-429 as biomarker for diagnosis, treatment and prognosis of cancers and its potential action mechanisms: A systematic literature review. Neoplasma 2020;67:215-28. [Crossref] [PubMed]
- Gu Y, Li X, Bi Y, et al. CCL14 is a prognostic biomarker and correlates with immune infiltrates in hepatocellular carcinoma. Aging (Albany NY) 2020;12:784-807. [Crossref] [PubMed]
- Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol 2019;12:133. [Crossref] [PubMed]
- Zhang T, Huang JW, Zeng Y. Surgery for patients with hepatocellular carcinoma and inferior vena cava tumor thrombus. Sichuan Da Xue Xue Bao Yi Xue Ban 2013;44:1005-8. [PubMed]
- Akateh C, Black SM, Conteh L, et al. Neoadjuvant and adjuvant treatment strategies for hepatocellular carcinoma. World J Gastroenterol 2019;25:3704-21. [Crossref] [PubMed]
- Zheng H, Pomyen Y, Hernandez MO, et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 2018;68:127-40. [Crossref] [PubMed]
- Fu Y, Liu S, Zeng S, et al. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J Exp Clin Cancer Res 2019;38:396. [Crossref] [PubMed]
- Lu C, Rong D, Zhang B, et al. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer 2019;18:130. [Crossref] [PubMed]
- Luo Q, Wang CQ, Yang LY, et al. FOXQ1/NDRG1 axis exacerbates hepatocellular carcinoma initiation via enhancing crosstalk between fibroblasts and tumor cells. Cancer Lett 2018;417:21-34. [Crossref] [PubMed]
- Lim CJ, Lee YH, Pan L, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut 2019;68:916-27. [Crossref] [PubMed]
- Arasada RR, Shilo K, Yamada T, et al. Notch3-dependent β-catenin signaling mediates EGFR TKI drug persistence in EGFR mutant NSCLC. Nat Commun 2018;9:3198. [Crossref] [PubMed]
- Thompson SM, Jondal DE, Butters KA, et al. Heat stress induced, ligand-independent MET and EGFR signalling in hepatocellular carcinoma. Int J Hyperthermia 2018;34:812-23. [Crossref] [PubMed]
- Tan KL, Pezzella F. Inhibition of NEDD8 and FAT10 ligase activities through the degrading enzyme NEDD8 ultimate buster 1: A potential anticancer approach. Oncol Lett 2016;12:4287-96. [Crossref] [PubMed]
- Liu X, Chen L, Ge J, et al. The Ubiquitin-like Protein FAT10 Stabilizes eEF1A1 Expression to Promote Tumor Proliferation in a Complex Manner. Cancer Res 2016;76:4897-907. [Crossref] [PubMed]
- Aichem A, Sailer C, Ryu S, et al. The ubiquitin-like modifier FAT10 interferes with SUMO activation. Nat Commun 2019;10:4452. [Crossref] [PubMed]
- Gao Y, Theng SS, Mah WC, et al. Silibinin down-regulates FAT10 and modulate TNF-α/IFN-γ-induced chromosomal instability and apoptosis sensitivity. Biol Open 2015;4:961-9. [Crossref] [PubMed]
- Bsibsi M, Peferoen LA, Holtman IR, et al. Demyelination during multiple sclerosis is associated with combined activation of microglia/macrophages by IFN-γ and alpha B-crystallin. Acta Neuropathol 2014;128:215-29. [Crossref] [PubMed]
- Liu L, Dong Z, Liang J, et al. As an independent prognostic factor, FAT10 promotes hepatitis B virus-related hepatocellular carcinoma progression via Akt/GSK3β pathway. Oncogene 2014;33:909-20. [Crossref] [PubMed]
- Yan J, Lei J, Chen L, et al. Human Leukocyte Antigen F Locus Adjacent Transcript 10 Overexpression Disturbs WISP1 Protein and mRNA Expression to Promote Hepatocellular Carcinoma Progression. Hepatology 2018;68:2268-84. [Crossref] [PubMed]
- Gao Y, Theng SS, Zhuo J, et al. FAT10, an ubiquitin-like protein, confers malignant properties in non-tumorigenic and tumorigenic cells. Carcinogenesis 2014;35:923-34. [Crossref] [PubMed]
- Lukasiak S, Schiller C, Oehlschlaeger P, et al. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene 2008;27:6068-74. [Crossref] [PubMed]
- Ren J, Wang Y, Gao Y, et al. FAT10 mediates the effect of TNF-α in inducing chromosomal instability. J Cell Sci 2011;124:3665-75. [Crossref] [PubMed]
- Choi Y, Kim JK, Yoo JY. NFκB and STAT3 synergistically activate the expression of FAT10, a gene counteracting the tumor suppressor p53. Mol Oncol 2014;8:642-55. [Crossref] [PubMed]
- Xiong H, Xiao H, Luo C, et al. GRP78 activates the Wnt/HOXB9 pathway to promote invasion and metastasis of hepatocellular carcinoma by chaperoning LRP6. Exp Cell Res 2019;383:111493 [Crossref] [PubMed]
- Zhang K, Chen L, Zhang Z, et al. Ubiquitin-like protein FAT10: A potential cardioprotective factor and novel therapeutic target in cancer. Clin Chim Acta 2020;510:802-11. [Crossref] [PubMed]
- Luo C, Xiong H, Chen L, et al. GRP78 Promotes Hepatocellular Carcinoma proliferation by increasing FAT10 expression through the NF-κB pathway. Exp Cell Res 2018;365:1-11. [Crossref] [PubMed]
- Ji F, Jin X, Jiao CH, et al. FAT10 level in human gastric cancer and its relation with mutant p53 level, lymph node metastasis and TNM staging. World J Gastroenterol 2009;15:2228-33. [Crossref] [PubMed]


