
Screening potential prognostic biomarkers for portal vein emboli in patients with hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer and the third most deadly cancer worldwide (1). Due to a lack of typical clinical symptoms or a complete absence of any symptoms in the early stages, most HCC patients are already in the middle and late stages of the disease at the time of diagnosis, and may even present with metastasis (2). The liver has a dual blood supply facilitated by the hepatic artery and portal vein. The anatomical structures of the arteries, veins, and bile ducts in the liver are complex. HCC is highly aggressive and heterogeneous, and is prone to intrahepatic metastasis and invasion of blood vessels. Portal vein tumor thrombus (PVTT) is the main form of hepatic vascular invasion and metastasis. Studies have shown that the incidence of PVTT in patients at initial diagnosis of HCC ranges from 10% to 62.2% (3-5). The formation of PVTT accelerates the progression of the patient’s disease course and induces portal hypertension, liver function deterioration, and other complications (6). The median overall survival (OS) of HCC patients with PVTT without any intervention is only 2.7 months (3). Indeed, PVTT is associated with the clinical stage of HCC and is an important factor for poor patient prognosis (6,7).
At present, there is no global unified guideline for the treatment of HCC patients with PVTT. Chinese guidelines recommend surgical resection, transcatheter chemoembolization, and systematic and symptomatic treatment. Multidisciplinary therapeutic assistance depends on liver function, tumor condition, and PVTT stage. The European Association for the Study of the Liver (EASL) and the National Comprehensive Cancer Network (NCCN) guidelines recommend targeted therapy and immunotherapy as first-line treatment for advanced HCC (7,8). As an inflammation-related tumor, HCC forms a complex immune tolerance microenvironment in the liver. Immunotherapy is therefore expected to be a promising treatment method for HCC (9).
Studies have shown that immune cells in the immune microenvironment contribute to the development of PVTT (10), but the specific mechanisms of action remain unclear. Thus, it is important to identify effective and sensitive immune-related molecular biomarkers for the diagnosis and treatment of patients with HCC complicated with PVTT. Wang et al. compared primary tumor (PT) and PVTT samples from HCC patients with matched adjacent normal tissue and identified 20 differentially expressed genes (DEGs) whose expression was significantly associated with OS and vascular invasion in HCC patients. Furthermore, these DEGs were found to modulate the aggressive phenotype of HCC cells (11). Other studies have shown that tumor-associated macrophages (TAM) are closely related to the expression of osteopontin, which is a key regulatory gene of HCC prognosis, angiogenesis, invasion, and metastasis (12-14). These reports confirm that immune cells in the immune microenvironment are closely related to the formation of PVTT and the prognosis of HCC patients.
To date, there have been few reports examining immunity and HCC complicated with PVTT. This current study used data from online bioinformatics databases to identify the DEGs, with a particular focus on differentially expressed immune-related genes, in PT and PVTT tissues. Bioinformatics analyses were performed to analyze the mRNA expression, biological function, immune invasion, and prognostic significance of the immune-related DEGs. The identification of novel immune biomarkers paves the way for improved diagnosis and the development of potential therapies for HCC patients complicated with PVTT.
We present the study in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-433).
Methods
Data sets and preprocessing
The GSE69164 and GSE77509 datasets were obtained from the Genome Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). There was a total of 22 samples in the GSE69164 dataset, including 11 cases of carcinoma and 11 paired cases of blood clots. The GSE77509 dataset had a total of 40 samples, including 20 cancers and 20 paired thrombus cases. The RNA-seq data for the HCC and normal samples (a total of 62 samples) were obtained from The Cancer Genome Atlas (TCGA; https://cancergenomes.nih.gov/) and normalized using the R package software.
Identification of the DEGs
Differential analysis was performed on all RNA-seq data, and the DEGs in the PT and paired PVTT samples were identified. The intersection of the DEGs and immune genes was used to obtain the differentially expressed immune-related genes.
Establishment of the prognostic model
To evaluate the relationship between the differentially expressed immune genes and patient prognosis, univariate Cox, Lasso, and multivariate Cox analyses were performed on the downloaded TCGA data. After identifying independent prognostic genes and determining their coefficients, the HCC patients were divided into a high-risk group and a low-risk group based on the median risk score coefficient. Receiver operating characteristic (ROC) curves were used to test the predictive power of the survival model.
Survival analysis
Univariate Cox regression analysis and the Kaplan-Meier method were used to evaluate the prognostic effect of the differentially expressed immune genes. When the Kaplan-Meier method was used to plot survival curves, a log-rank P<0.05 was considered statistically significant.
Tumor-infiltrating immune cell (TIC) analysis
The single-sample Gene Set Enrichment Analysis (ssGSEA) method was used to calculate the content of 28 types of HCC immune cells in the TCGA cohort, and the correlation between gene expression and immune cell expression was analyzed.
Enrichment function analysis and protein-protein interaction (PPI) network construction
Using the clusterProfiler, enrichplot, and ggplot2 packages, the potential biological functions and pathways of the DEGs in HCC were explored using R language. The clusterProfiler and enrichplot packages use hypergeometric distributions to calculate each item of the P and Q values for Gene Ontology (GO) or Kyoto Encyclopedia of Genes and Genomes (KEGG). AP and Q values less than 0.05 were considered significant enrichment functions. The PPI network of different genes was constructed through the string website, and the top 10 genes were calculated by Cytohubba plug-in using Cytoscape software.
Drug sensitivity analysis
The HCC gene expression matrix of TCGA was used to calculate the sensitivity of the samples to the drug based on the amount of gene expression in each sample. The difference between the high-risk group and the low-risk group was compared using the rank-sum test to determine the sensitivity of the drug.
Statistical analysis
Wilcoxon rank-sum test was used to compare the differences in gene expression between tumor tissues and carcinoma tissues. The Kaplan-Meier method was used to draw survival curves, and the log-rank test was performed to test its significance. The ssGSEA algorithm was used to evaluate TICs. All statistical data were completed using the R language software package (https://www.r-project.org/), and P<0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Identification of DEGs and differentially expressed immune genes in PT and PVTT
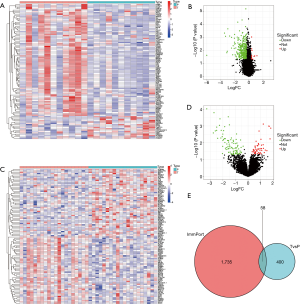
The GSE69164 and GSE77509 datasets were used to screen the DEGs in the PT and PVTT samples. A total of 458 DEGs were associated with tumor thrombi. When these DEGs were intersected with 1735 immune genes, a total of 58 immune-related DEGs were identified (Figure 1).
Construction and validation of a prognostic model
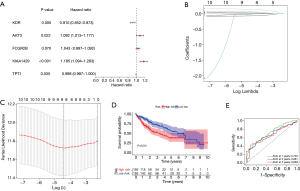
To further understand the prognostic value of the 58 differentially expressed immune genes, univariate and multivariate analyses were performed. A total of five predictive genes, KDR, AKT3, FCGR2B, KIAA1429, and TPT1, were identified (P<0.05). The risk score formula for OS prediction was as follows: risk score = (0.088 × AKT3) + (0.042 × FCGR2B) + (0.170 × KIAA1429) + (0.002 × TPT1) + (0.094 × KDR). Patients were divided into a high-risk group (n=185) and a low-risk group (n=185) based on the median risk score. Survival analysis showed that patients with a high-risk score generally had poorer prognosis compared to patient with a low-risk score (P<0.001; Figure 2A-2D). In addition, time-dependent ROCs were used to evaluate the prognostic value of the model. The results showed that the area under the ROC curve (AUC) of 1-, 3-, and 5-year OS was 0.704, 0.661, and 0.659, respectively, suggesting that the model has a good predictive ability for 1-year OS (Figure 2E). Subsequently, the expression of these five genes and patient prognosis was examined in HCC tumor tissues and normal tissues. The results demonstrated that there was no significant difference in AKT3 and KDR expression between tumor tissues and normal tissues. However, patients with high KDR expression had a better prognosis (P=0.026). The expression of FCGR2B was significantly lower in tumor tissues compared to normal tissues, but its expression was not correlated with patient prognosis (P=0.82). Both KIAA1429 and TPT1 were significantly overexpressed in tumor tissues. Patients with low KIAA1429 expression and high TPT1 expression had better prognosis (P=0.017; Figure 3). HCC is highly heterogeneous, and this may be related to the evolution and heterogeneity of the tumor.
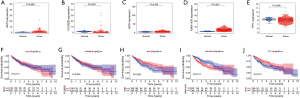
Immunoinfiltration analysis
The HCC cohort data from TCGA were used to analyze the level of immune infiltration. Except for central memory CD4 T cells and memory B cells, the other 26 types of immune cells showed different degrees of immune infiltration in HCC (Figure 4A), and HCC was closely related to immune cells. Further analysis showed that FCGR2B, KDR, and AKT3 were positively correlated with the expression of most immune cells, while KIAA1429 was negatively correlated with the expression of most immune cells. In addition, TPT1 was mainly positively correlated with the high expression of immune cells. However, it was negatively correlated with CD56dim natural killer cells and central memory CD8 T cell (Figure 4B).

Rich set function analysis and PPI network construction
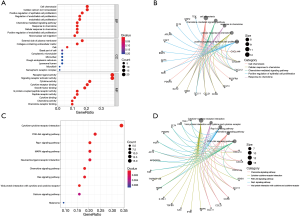
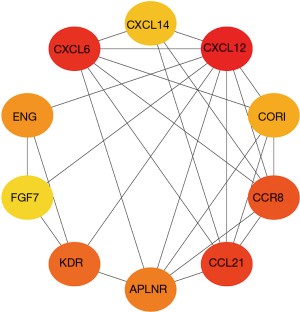
The potential function of the DEGs identified in the Venn diagram was further investigated using GO and KEGG pathway enrichment analysis. The enrichment function analysis revealed that the top three biological processes (BP) were cell chemotaxis, cellular calcium homeostasis, and positive regulation of epithelial cell proliferation. The top three cellular components (CC) were external side of the plasma membrane, collagen-containing extracellular matrix, and basal part of the cell. The identified molecular functions (MF) included receptor-ligand activity, signaling receptor activator activity, and cytokine activity. KEGG signaling pathway analysis showed that the DEGs were mainly associated with the MAPK signaling pathway, the RAP1 signaling pathway, the PI3K-Akt signaling pathway, and cytokine-cytokine receptor interaction (Figure 5). The relationship between DEGs was screened and the PPI network was constructed (Figure 6). These results indicated that the screened DEGs have a certain role in the immune system.
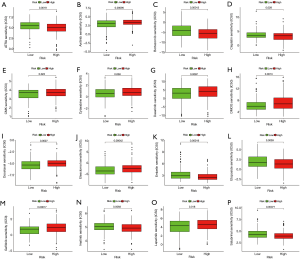
Drug sensitivity analysis
To examine the relationship between the risk scores and sensitivity, the TCGA data was analyzed. The genes identified were highly sensitive to certain drugs including all-trans retinoic acid (P=0.0019), axitinib (P=0.00095), gefitinib (P=0.00017), imatinib (P=0.0093), and lapatinib (P=0.018) (Figure 7). These results suggested that the model can not only predict the prognosis of patients but also assist in the clinical treatment decision-making process.
Discussion
The prognosis of HCC patients complicated with PVTT is extremely poor, and to date, there is no specific sensitive marker that can predict the formation of PVTT and the prognosis of patients. Studies have shown that serum S100p is overexpressed in HCC and is highly correlated with the formation of PVTT and microvascular invasion (MVI). Thus, S100p may represent a novel differential diagnostic marker for HCC and may serve as a potential predictor of preoperative MVI status (15). Xu et al. demonstrated that the levels of serum SCAND3 methylation is an independent risk factor for predicting PVTT in HCC patients (16). Liu et al. showed that microRNA-135a promoted the formation of PVTT in nude mice by inhibiting metastasis suppressor 1, suggesting that microRNA-135a may be a potential therapeutic target for the treatment of PVTT at the molecular level (17). However, to date, there have been few studies examining the DEGs in PT and PVTT tissues. Compared with adjacent normal liver tissue, HCC with PVTT show significant molecular changes. Differential expression of some genes can regulate the invasion of HCC cells. Gene analysis between HCC and PVTT also suggested that extracellular matrix receptor interactions are associated with venous invasion of HCC (11,18). Studies have shown that the circular RNA circ0003998 is highly expressed in PVTT and PT tissues and is associated with invasive characteristics. Further in vitro and in vivo experiments confirmed that circ0003998 promotes the epithelial-mesenchymal transformation of HCC (19). This current study identified a large number of DEGs in PT and PVTT samples, suggesting that these genes may be potential prognostic markers in HCC patients with PVTT.
Studies have shown that KDR is the main receptor that promotes the pro-angiogenesis of vascular endothelial growth factor (VEGF), and KDR/FLK-1 is one of the main receptors of VEGF. Treatment targeting KDR/FLK-1 can inhibit tumor angiogenesis, increase tumor cell proliferation, and inhibit the development of HCC (20). In addition, studies have shown that KDR can serve as an independent prognostic biomarker in patients with unresectable HCC (21). AKT3 is an adverse prognostic factor in patients with HCC (22), and activation of AKT3 promotes HCC stem cell properties and reduces chemosensitivity (23). In addition, KIAA1429 is significantly overexpressed in HCC tissues and is associated with poor prognosis. Inhibition of KIAA1429 impedes tumor cell proliferation and metastasis in vitro and in vivo (24). Our current study identified the differentially expressed immune genes, KDR, AKT3, FCGR2B, KIAA1429, and TPT1, all of which are correlated with the prognosis of HCC patients with PVTT. Furthermore, the model constructed showed a certain predictive ability for patient prognosis.
HCC is a typical inflammatory-related malignancy, and its microenvironment contains a large number of macrophages as well as innate immune and adaptive cells, forming a complex immune tolerance microenvironment (25,26). In this study, a significant degree of immune cell infiltration was observed in HCC tissue. Furthermore, the differentially expressed immune genes FCGR2B, KDR, and AKT3 were mainly positively correlated with the expression of immune cells, while KIAA1429 was negatively correlated with the expression of immune cells. Interestingly, TPT1 was positively correlated with certain types of immune cells, and negatively correlated with other immune cells. A growing number of studies have shown that TAM affect the angiogenesis, invasion, and metastasis of HCC, as well as patient prognosis and tumor progression (13,27-29). In addition, increased numbers of T regulatory (Treg) cells in HCC tissues is associated with PVTT formation, metastasis and recurrence, and poor prognosis (30). Immune cells can also interact with each other. Tumor-associated neutrophils can recruit TAM cells and Treg cells to promote HCC growth and angiogenesis (31). To further understand the function of the differentially expressed immune genes, GO and KEGG analyses were performed. The results demonstrated that the DEGs were mainly involved in cytokine-cytokine receptor interaction, the MAPK signaling pathway, and the PI3K-Akt signaling pathway. The PI3K and MAPK signaling pathways rely on cells that adhere to the external matrix via integrins, which are molecules that may amplify these signals (32). Further PPI analysis suggested that the DEGs may be involved in the regulation of various non-cellular components such as inflammatory factors and chemokines. Recent studies have found that CXCL12 and its receptors, CXCR7 and CXCR4, promote invasion and metastasis of HCC, and are closely related to tumor progression, neovascularization, epithelial-mesenchymal transformation, and poor prognosis of HCC (33,34). Therefore, the DEGs identified in this study may be involved in the regulation of immune infiltration, and thereby regulate the progression of HCC.
In recent years, the emergence of immunotherapy has provided a new option for the treatment of HCC. However, the efficacy of immunotherapy can be limited by low objective response rates, increased incidence of adverse reactions, and drug resistance. This current study demonstrated that the differentially expressed immune genes identified were highly sensitive to atrt, axitinib, gefitinib, imatinib, and lapatinib. A randomized phase II clinical study of second-line treatment for advanced HCC demonstrated that axitinib was safe and significantly improved the progression-free survival, the time to tumor progression, and the clinical benefit rates in a subset of the Asian population (35). Lapatinib has also been shown to be well tolerated in a multi-institution phase II clinical study in patients with advanced HCC. However, the molecular and clinical features of the patients were not fully defined (36). Additional clinical trials are warranted to confirm the potential efficacy of axitinib and lapatinib in the treatment of patients with HCC complicated with PVTT.
There were some limitations to this present investigation. First, the small number of cases in the study may affect the accuracy and reliability of the results. Second, TCGA data mainly relates to American patients, and this may lead to selection bias. Expanded case numbers and basic studies in different populations are needed to verify our results.
Conclusions
In conclusion, this study identified a large number of DEGs in the PT and PVTT of HCC patients. The differentially expressed immune genes identified may promote the progression of PVTT by participating in the regulation of non-cellular components such as the extracellular matrix, inflammatory factors, and chemokines. Specifically, the differentially expressed immune genes KDR, AKT3, FCGR2B, KIAA1429, and TPT1 may be potential predictors and therapeutic targets for HCC patients with PVTT. Further basic studies and functional experiments are warranted to confirm these results.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-433
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-433). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Duran SR, Jaquiss RDB. Hepatocellular carcinoma. N Engl J Med 2019;381:e2 [Crossref] [PubMed]
- Zhang ZM, Lai EC, Zhang C, et al. The strategies for treating primary hepatocellular carcinoma with portal vein tumor thrombus. Int J Surg 2015;20:8-16. [Crossref] [PubMed]
- Lu J, Zhang XP, Zhong BY, et al. Management of patients with hepatocellular carcinoma and portal vein tumour thrombosis: comparing east and west. Lancet Gastroenterol Hepatol 2019;4:721-30. [Crossref] [PubMed]
- Cheng S, Chen M, Cai J, et al. Chinese expert consensus on multidisciplinary diagnosis and treatment of hepatocellular carcinoma with portal vein tumor thrombus (2018 edition). Liver Cancer 2020;9:28-40. [Crossref] [PubMed]
- Chan SL, Chong CC, Chan AW, et al. Management of hepatocellular carcinoma with portal vein tumor thrombosis: review and update at 2016. World J Gastroenterol 2016;22:7289-300. [Crossref] [PubMed]
- European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu; European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182-236. [Crossref]
- Benson AB, D'Angelica MI, Abbott DE, et al. Hepatobiliary cancers, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021;19:541-65. [Crossref] [PubMed]
- Johnston MP, Khakoo SI. Immunotherapy for hepatocellular carcinoma: current and future. World J Gastroenterol 2019;25:2977-89. [Crossref] [PubMed]
- Brodt P. Role of the microenvironment in liver metastasis: from pre- to prometastatic niches. Clin Cancer Res 2016;22:5971-82. [Crossref] [PubMed]
- Wang D, Zhu Y, Tang J, et al. Integrative molecular analysis of metastatic hepatocellular carcinoma. BMC Med Genomics 2019;12:164. [Crossref] [PubMed]
- Zhu W, Guo L, Zhang B, et al. Combination of osteopontin with peritumoral infiltrating macrophages is associated with poor prognosis of early-stage hepatocellular carcinoma after curative resection. Ann Surg Oncol 2014;21:1304-13. [Crossref] [PubMed]
- Dong P, Ma L, Liu L, et al. CD86+/CD206+, diametrically polarized tumor-associated macrophages, predict hepatocellular carcinoma patient prognosis. Int J Mol Sci 2016;17:320. [Crossref] [PubMed]
- Chen F, Chen J, Yang L, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol 2019;21:498-510. [Crossref] [PubMed]
- Qi LN, Ma L, Wu FX, et al. S100P as a novel biomarker of microvascular invasion and portal vein tumor thrombus in hepatocellular carcinoma. Hepatol Int 2021;15:114-26. [Crossref] [PubMed]
- Xu F, Zhang L, Xu Y, et al. Hypermethylation of SCAND3 and Myo1g Gene Are Potential Diagnostic Biomarkers for Hepatocellular Carcinoma. Cancers (Basel) 2020;12:2332. [Crossref] [PubMed]
- Liu S, Guo W, Shi J, et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol 2012;56:389-96. [Crossref] [PubMed]
- Zhang H, Ye J, Weng X, et al. Comparative transcriptome analysis reveals that the extracellular matrix receptor interaction contributes to the venous metastases of hepatocellular carcinoma. Cancer Genet 2015;208:482-91. [Crossref] [PubMed]
- Song LN, Qiao GL, Yu J, et al. Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J Exp Clin Cancer Res 2020;39:114. [Crossref] [PubMed]
- Yoshiji H, Kuriyama S, Hicklin DJ, et al. KDR/Flk-1 is a major regulator of vascular endothelial growth factor-induced tumor development and angiogenesis in murine hepatocellular carcinoma cells. Hepatology 1999;30:1179-86. [Crossref] [PubMed]
- Machado MV, Janeiro A, Miltenberger-Miltenyi G, et al. Genetic polymorphisms of proangiogenic factors seem to favor hepatocellular carcinoma development in alcoholic cirrhosis. Eur J Gastroenterol Hepatol 2014;26:438-43. [Crossref] [PubMed]
- Zhang Y, Guo X, Yang M, et al. Identification of AKT kinases as unfavorable prognostic factors for hepatocellular carcinoma by a combination of expression profile, interaction network analysis and clinical validation. Mol Biosyst 2014;10:215-22. [Crossref] [PubMed]
- Shu G, Su H, Wang Z, et al. LINC00680 enhances hepatocellular carcinoma stemness behavior and chemoresistance by sponging miR-568 to upregulate AKT3. J Exp Clin Cancer Res 2021;40:45. [Crossref] [PubMed]
- Lan T, Li H, Zhang D, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer 2019;18:186. [Crossref] [PubMed]
- Nishida N, Kudo M. Immunological microenvironment of hepatocellular carcinoma and its clinical implication. Oncology 2017;92:40-9. [Crossref] [PubMed]
- Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025-41. [Crossref] [PubMed]
- Galdiero MR, Bonavita E, Barajon I, et al. Tumor associated macrophages and neutrophils in cancer. Immunobiology 2013;218:1402-10. [Crossref] [PubMed]
- Wenes M, Shang M, Di Matteo M, et al. Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab 2016;24:701-15. [Crossref] [PubMed]
- Neophytou CM, Pierides C, Christodoulou MI, et al. The role of tumor-associated myeloid cells in modulating cancer therapy. Front Oncol 2020;10:899. [Crossref] [PubMed]
- Huang Y, Wang FM, Wang T, et al. Tumor-infiltrating FoxP3+ Tregs and CD8+ T cells affect the prognosis of hepatocellular carcinoma patients. Digestion 2012;86:329-37. [Crossref] [PubMed]
- Zhou SL, Zhou ZJ, Hu ZQ, et al. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology 2016;150:1646-58.e17. [Crossref] [PubMed]
- Filliol A, Schwabe RF. Contributions of fibroblasts, extracellular matrix, stiffness, and mechanosensing to hepatocarcinogenesis. Semin Liver Dis 2019;39:315-33. [Crossref] [PubMed]
- Lin L, Han MM, Wang F, et al. CXCR7 stimulates MAPK signaling to regulate hepatocellular carcinoma progression. Cell Death Dis 2014;5:e1488 [Crossref] [PubMed]
- Yang J, Zhang L, Jiang Z, et al. TCF12 promotes the tumorigenesis and metastasis of hepatocellular carcinoma via upregulation of CXCR4 expression. Theranostics 2019;9:5810-27. [Crossref] [PubMed]
- Kang YK, Yau T, Park JW, et al. Randomized phase II study of axitinib versus placebo plus best supportive care in second-line treatment of advanced hepatocellular carcinoma. Ann Oncol 2015;26:2457-63. [Crossref] [PubMed]
- Bekaii-Saab T, Markowitz J, Prescott N, et al. A multi-institutional phase II study of the efficacy and tolerability of lapatinib in patients with advanced hepatocellular carcinomas. Clin Cancer Res 2009;15:5895-901. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


