
Effects of YAP1 and SFRP2 overexpression on the biological behavior of colorectal cancer cells and their molecular mechanisms
Introduction
Colorectal cancer (CRC) is one of the most common malignancies in the world, and has a high morbidity and mortality rate (1,2). CRC is caused by multiple factors and multiple mechanisms, which are associated with abnormal cell signaling pathways, such as the Hippo/Yes-associated protein 1 (YAP1) pathway and the Wnt/β-catenin pathway (3,4). In the Hippo/YAP1 signaling pathway, the activation of YAP1 is associated with tumor acquisition of malignant characteristics, including anti-cancer therapy resistance, the metastasis of cancer stem cells, and epithelial-mesenchymal transformation (5-7). In addition, YAP1 has been shown to be associated with poor prognosis and reduced survival in many human cancers (8-10). Thus, YAP1 can be used as a marker of poor prognosis and drug resistance in cancers. In the Wnt/β-catenin signaling pathway, β-catenin can lead to tumor recurrence and treatment resistance (11,12). Secreted frizzled-related proteins (SFRPs) are a type of Wnt signaling inhibitor that are expressed at significantly reduced levels in a variety of cancers and are associated with poor disease progression and prognosis (13,14). SFRPs can be used as non-invasive biomarkers for the early diagnosis of CRC (15).
In the present study, we investigated correlations among the expression of YAP1, SFRP2, and β-catenin, and the effects of YAP1 and SFRP2 on the proliferation and apoptosis of CRC cells. The aim of the study was to elucidate the mechanism underlying CRC development and progression and provide novel strategies for its treatment. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-418).
Methods
Promoter screening
SW480 cells (BeNa Culture Collection, Beijing, China) in the logarithmic growth phase were seeded into 24 well plates. The cells were cultured to ~70% confluence, and then transfected with 6 promoter plasmids (ZHBY Biotech Co., Ltd, Jiangxi, China) using Lipofectamine™ 3000 Transfection Reagent (Invitrogen™, USA) in accordance with the manufacturer’s protocol. Fresh medium was added to each well after transfection for 6 hours. Fluorescence signals were observed after transfection for 48 hours.
Lentiviruses packages and infects cells
The gene expression vectors of YAP1 and SFRP2 were constructed using the screened promoters and packaged with lentivirus (ZHBY Biotech Co., Ltd, Jiangxi, China). Subsequently, the multiplicity of infection (MOI) values of different gradients were set to select the MOI values with the best cell status and the strongest fluorescence expression after lentivirus infection. After the cells were infected with the optimal concentration of lentivirus, the overexpression efficiency was verified by quantitative polymerase chain reaction (qPCR) and Western blot.
qPCR
Total ribonucleic acid (RNA) was extracted from the CRC cells using TRIzon Reagent (CWBIO, Beijing, China) in accordance with the manufacturer’s instructions. To synthesize the complementary deoxyribonucleic acid (cDNA), reverse transcription (RT) was performed using HiScript II Q RT Super Mix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China). The aliquots of cDNA were amplified using 2× SYBR Green PCR Master Mix (Lifeint, Xiamen, China). β-actin was used as an internal control. The relative messenger RNA (mRNA) expressions were quantified using the 2−ΔΔCt method. IBM SPSS Statistics 20.0 was used to test the relative expression levels of each group and evaluate the relative expression differences between the experimental group and the control group. The following primers were used: YAP1 forward 5'-GCT TGA CCC TCG TTT TGC CAT-3' and reverse 5'-GAA TTT GCT GTG CTG GGA TTG ATA-3'; SFRP2 forward 5'-ACC TAG ACG AGA CCA TCC AG-3' and reverse 5'-ATA CCT TTG GAG CTT CCT CG-3'; β-actin forward 5'-TGG CAC CCA GCA CAA TGA A-3' and reverse 5'-CTA AGT CAT AGT CCG CCT AGA AGC A-3'.
Western blot analysis
The cells were lysed using RIPA (Radio Immunoprecipitation Assay) buffer (Apply Gen, Beijing, China). The lysis solution was collected after centrifugation at 12,000 ×g for 10 minutes at 4 °C. The protein concentration was determined using a BCA (Bicinchoninic Acid) Protein Assay Kit (CWBIO, Beijing, China). Total proteins were loaded in 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Beijing, China). The membranes were blocked with 3% skim milk (Apply Gen, Beijing, China) for 1 hour, and then incubated with primary antibody overnight. After washing, the membranes were incubated with a secondary antibody for 2 hours at room temperature. The immunoblots were acquired using the SuperSignal® West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, RJ239676) in accordance with the manufacturer’s protocol. The following antibodies were used: Rabbit Anti-YAP1 (1:2,000; Protein Tech Group, Inc., Chicago, IL, USA), Rabbit Anti-β-catenin (1:2,000; Abcam, Cambridge, MA, USA), Mouse Anti-SFRP2 (1:1,000; Proteinab Biotech Co., Ltd, China) and Mouse Anti-GAPDH (1:2,000; ZSGB-BIO, China). Goat Anti-Rabbit and Anti-Mouse Immunoglobulin Gs (IgGs) (H+L) (ZSGB-BIO, China) were used as secondary antibodies diluted at 1:2,000.
Co-immunoprecipitation assay
For the co-immunoprecipitation assay, we used the Pierce™ Classic Magnetic IP/Complex Immunoprecipitation (Co-IP) Kit (Thermo Fisher Scientific,88804) in accordance with manufacturer’s protocol. 200 µL of cell lysate was incubated with a mixture of 20 µL of protein A/G beads and 5 µL of conjugated antibody to YAP1/SFRP2 at 4 °C overnight with rotation. The beads were then washed with IP (Immunoprecipitation) lysis/wash buffer and PBS. 200 µL of elution buffer was added to the beads, which were incubated at room temperature and mixed for 5–10 minutes. The supernatant was transferred to a new tube. Finally, the samples were loaded onto a SDS-PAGE gel for a western blot analysis.
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde and blocked with 5% BSA. The cells were then incubated with a mixture of 2 primary antibodies (Rabbit Anti-YAP1, 1:200 and Mouse Anti-SFRP2, 1:100/1:200) at 4 °C overnight. The cells grown on coverslips were washed with PBS and incubated with a fluorescent labeled secondary antibody (Goat Anti-Rabbit and Anti-Mouse IgG, 1:100, ABclonal Technology Co., Ltd., Wuhan, China) in the dark for 30 minutes at 37 °C. The nuclei were counterstained by 4’,6- diamidino-2-phenylindole (DAPI, Key GEN BioTECH Co., Ltd, Jiangsu, China). Images were captured using a fluorescence microscope (OLYMPUS, CKX53).
Cell viability assay
CRC cells were seeded in 96-well culture plates (Corning, Shanghai, China). A total of 10 µL CCK8 reagent (KeyGEN BioTECH Co., Ltd, China) was added to each well and incubated for 4 hours. The absorbance was measured at 450 nm using a microplate reader.
Flow cytometry assay
The CRC cells were washed with PBS and collected after centrifugation at 1,500 ×g for 3 minutes. The cells were remixed with 500 µL ice-cold 1× binding buffer. Then, 5 µL of Annexin V-APC and 10 µL of 7-AAD (Multi Sciences Co., Ltd, Hangzhou, China) were added to each tube, and the cells were incubated in the dark for 10 minutes at room temperature. Cell apoptosis was detected by NovoCyte™ flow cytometry (ACEA BIO Co., Ltd, Hangzhou, China).
Statistical analysis
All statistical analyses were performed in IBM SPSS Statistics (Version 20.0, IBM, Armonk, NY, USA). The quantitative results are expressed as mean ± standard deviation (
Results
Promoter screening
To construct the vectors, we first performed promoter screening. We selected the following 6 plasmids: pLVX-CMV-IRES-EGFP-puro, pLVX-EF1α-IRES -EGFP-puro, pLVX-SV40-IRES-EGFP-puro, pLVX-PGK-IRES-EGFP-puro, pLVX-CMV-IRES-ZsGreen, and pLVX-EF1α-IRES-EGFP-PGK-puro. The fluorescence expression level of pLVX-EF1α-IRES-EGFP-PGK-puro in SW480 cells was significantly higher than that of the others (see Figure 1). As pLVX- EF1α-IRES-EGFP-PGK-puro exhibited a higher fluorescence expression than the other 6 plasmids, it was selected to perform the vector construction.
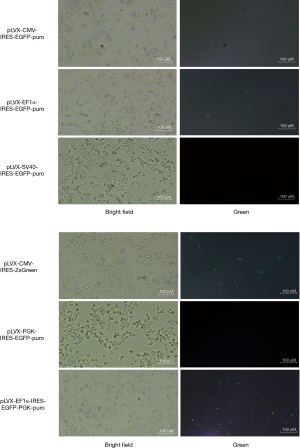
The construction of overexpressed cell lines
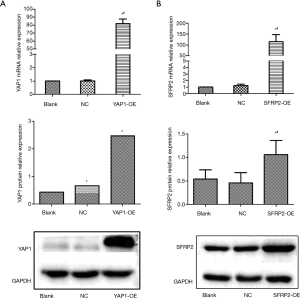
To verify the efficiency overexpression of YAP1 and SFRP2 in SW480 cells, we detected the expression levels of YAP1 and SFRP2 by qPCR and Western blot. In the CRC cells, the expression levels of YAP1 overexpression groups (see Figure 2A) and SFRP2 overexpression groups (see Figure 2B) were significantly higher than Blank and NC groups. Thus, we constructed the overexpression cell lines of YAP1 and SFRP2.
The overexpression of YAP1 had no effect on the expression of SFRP2
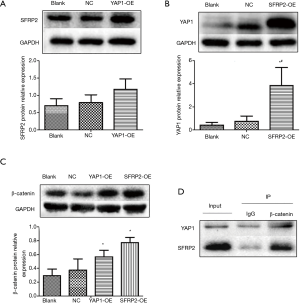
A Western blot analysis was performed to determine the effects of YAP1 on SFRP2 expression at the protein level. The overexpression of YAP1 had no effect on the expression of SFRP2 in CRC cells (see Figure 3A). The results suggest that YAP1 is not an upstream regulator of SFRP2 in CRC.
The overexpression of SFRP2 promotes the expression of YAP1 in CRC cells
A Western blot analysis was performed to determine the effects of SFRP2 on YAP1 expression at the protein level. The expression of YAP1 was upregulated in SFRP2 overexpression group (see Figure 3B). The results suggest that SFRP2 may be an upstream regulator of YAP1 in CRC.
The overexpression of YAP1 and SFRP2 promotes the expression of β-catenin in CRC cells
A Western blot analysis was used to detect the expression of β-catenin protein in each group. The results showed that the relative expression of β-catenin protein in YAP1 and SFRP2 overexpression groups was significantly higher than that in the Blank group and NC group (see Figure 3C).
Relationship between YAP1, SFRP2 expression and β-catenin expression
A co-immunoprecipitation assay was used to detect the interaction between YAP1, SFRP2 expression, and β-catenin expression. The results showed that both YAP1 and SFRP2 interacted with β-catenin expression (see Figure 3D).
The localization and expression of YAP1 in SW480 cells
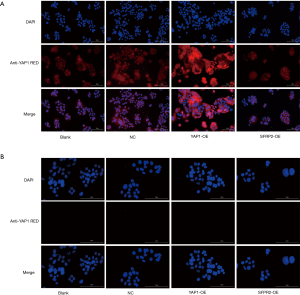
The expression of YAP1 and SFRP2 in each group was detected by immunofluorescence. The results showed that YAP1 was expressed in both the cytoplasm and nucleus of each group, and the relative expression of the YAP1 overexpressed group was the highest (see Figure 4A). However, there was no expression of SFRP2 in each group (see Figure 4B).
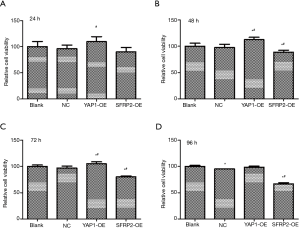
The effects of YAP1 and SFRP2 overexpression on the proliferation of CRC cells
To investigate the effects of YAP1 and SFRP2 on the proliferation behavior of CRC cells in vitro, we performed Cell Counting Kit-8 (CCK8) assays. Our results showed that the overexpression of YAP1 promoted the proliferation of SW480 cells (see Figure 5). Conversely, the overexpression of SFRP2 suppressed the proliferation of SW480 cells (see Figure 5).
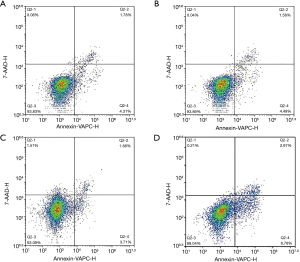
The effects of YAP1 and SFRP2 overexpression on the apoptosis of CRC cells
To explore the effects of YAP1 and SFRP2 on CRC cell apoptosis, we performed a flow cytometry assay. Our results showed that CRC cells with SFRP2 overexpression had a significantly reduced apoptosis capacity compared to NC (see Figure 6). Conversely, there was no effect on CRC cells when YAP1 was overexpressed (see Figure 6).
Discussion
CRC is one of the most common malignancies in the world, and has a high fatality rate (16). At present, the molecular basis of colorectal tumor formation is still not fully understood. In recent years, with the development of modern molecular biology, the exploration of the molecular mechanism and targeted therapy of CRC at the gene level has become a popular area of research. YAP1 is a powerful regulator of tumor cell proliferation, and is closely related to the formation and development of a variety of tumors (17,18). Many YAP1/TEAD (TEA domain family member) target genes are also associated with tumor progression, including baculoviral IAP repeat containing 5 (BIRC5), cyclin D1 (CCND1), and forkhead box M1 (FOXM1) (6,17,18). Of these, FOXM1 is highly expressed in a variety of human cancer cells (6,17,18). The Wnt signaling pathway is one of the key cascades in regulating organ development and stem cell properties and is closely associated with cancer, most notably CRC (19). SFRPs inhibit the abnormal activation of the Wnt pathway and inhibit cell proliferation (20,21). Thus, research that examines how YAP1 and SFRP2 are involved in the molecular mechanisms and biological behaviors of CRC cells may provide new evidence of novel diagnostic and prognostic biomarkers and therapeutic targets for CRC.
In the present study, we found a correlation between YAP1, SFRP2, and β-catenin expression in CRC cell lines. We found that the overexpression of SFRP2 promotes the expression of YAP1 and the overexpression of YAP1 and SFRP2 promote the expression of β-catenin in CRC cells. Both YAP1 and SFRP2 interacted with the β-catenin protein. It has been reported that downregulating the expression of YAP1 can suppress the expression of β-catenin (22), and that YAP1 and β-catenin interact with each other (23). According to one report, the elevated expression of SFRP2 increases the expression of β-catenin (24). In the present study, we found that YAP1 protein was expressed in the cytoplasm and nucleus of each group. Huang et al.’s report confirmed these results (25). However, the immunofluorescence assay showed no expression of SFRP2 in the cells of each group. This may have been due to the low content of SFRP2 protein, the low number of cells expressing SFRP2 protein, or the antibody failing to meet the level of specificity required for the immunofluorescence test. We intend conduct subsequent experiments with an optimized scheme in the future. However, our findings indicate that there may be an interaction between the Hippo/YAP1 pathway and the Wnt/β-catenin pathway in CRC. The mechanism of pathway crossover needs to be studied, as it could have great significance to cancer treatments and interventions (26).
To determine the potential function of YAP1 and SFRP2 in CRC development and progression, we analyzed the cell proliferation and apoptosis of overexpressed YAP1 and SFRP2. We found that the overexpression of YAP1 promoted CRC cell proliferation, but the overexpression of SFRP2 suppressed CRC cell proliferation and promoted apoptosis. It has been reported that the overexpression of SFRP1 and SFRP2 inhibited cell proliferation (27-29). The overexpression of YAP1 promoted proliferation and inhibited apoptosis in the SW480 and HCT116 cells (30). However, in this study, YAP1 overexpression did not significantly inhibit apoptosis, and we are of the view that other unknown regulatory mechanisms may limit the role of YAP1. These results indicated that YAP1 may act as a tumor activator and SFRP2 may act as a tumor inhibitor. SFRP2, which is a member of the SFRP family of proteins, inhibits the Wnt pathway to limit cell proliferation. It has been reported that the expression level of SFRPs was downregulated in various types of cancer (30). Serum SFRP2 hypermethylation may be a non-invasive biomarker for CRC screening and a prognostic predictor for postoperative CRC patients (31,32).
In conclusion, there may be an interaction between the Hippo/YAP1 pathway and the Wnt/β-catenin pathway in CRC. YAP1 and SFRP2 are involved in the formation and development of CRC. These findings indicate that YAP1 or SFRP2 may be promising targets for CRC treatments in the future.
Acknowledgments
Funding: This study was supported by the Natural Science Foundation of Anhui Province (1808085MH283) and the Doctoral Research Funded Project of the Anhui Medical University (XJ201614).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-418
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-418
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-418). All authors have reported that this work has received funding from the Natural Science Foundation of Anhui Province (1808085MH283) and the Doctoral Research Funded Project of the Anhui Medical University (XJ201614). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by Fuyang Hospital of Anhui Medical University (No. KY2021015). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ahmed M. Colon Cancer: A Clinician's Perspective in 2019. Gastroenterology Res 2020;13:1-10. [Crossref] [PubMed]
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [Crossref] [PubMed]
- Lee SK, Hwang JH, Choi KY. Interaction of the Wnt/β-catenin and RAS-ERK pathways involving co-stabilization of both β-catenin and RAS plays important roles in the colorectal tumorigenesis. Adv Biol Regul 2018;68:46-54. [Crossref] [PubMed]
- Farooqi AA, de la Roche M, Djamgoz MBA, et al. Overview of the oncogenic signaling pathways in colorectal cancer: Mechanistic insights. Semin Cancer Biol 2019;58:65-79. [Crossref] [PubMed]
- Segrelles C, Paramio JM, Lorz C. The transcriptional co-activator YAP: A new player in head and neck cancer. Oral Oncol 2018;86:25-32. [Crossref] [PubMed]
- Nilsson MB, Sun H, Robichaux J, et al. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med 2020;12:eaaz4589 [Crossref] [PubMed]
- Guimei M, Alrouh S, Saber-Ayad M, et al. Inhibition of Yes-Associated Protein-1 (YAP1) Enhances the Response of Invasive Breast Cancer Cells to the Standard Therapy. Breast Cancer (Dove Med Press) 2020;12:189-99. [Crossref] [PubMed]
- Guo L, Chen Y, Luo J, et al. YAP1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio 2019;9:437-45. [Crossref] [PubMed]
- Muhammad JS, Guimei M, Jayakumar MN, et al. Estrogen-induced hypomethylation and overexpression of YAP1 facilitate breast cancer cell growth and survival. Neoplasia 2021;23:68-79. [Crossref] [PubMed]
- Godlewski J, Kiezun J, Krazinski BE, et al. The Immunoexpression of YAP1 and LATS1 Proteins in Clear Cell Renal Cell Carcinoma: Impact on Patients' Survival. Biomed Res Int 2018;2018:2653623 [Crossref] [PubMed]
- Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev 2018;62:50-60. [Crossref] [PubMed]
- Uppada SB, Gowrikumar S, Ahmad R, et al. MASTL induces Colon Cancer progression and Chemoresistance by promoting Wnt/β-catenin signaling. Mol Cancer 2018;17:111. [Crossref] [PubMed]
- Deshmukh A, Arfuso F, Newsholme P, et al. Epigenetic demethylation of sFRPs, with emphasis on sFRP4 activation, leading to Wnt signalling suppression and histone modifications in breast, prostate, and ovary cancer stem cells. Int J Biochem Cell Biol 2019;109:23-32. [Crossref] [PubMed]
- Yin P, Wang W, Zhang Z, et al. Wnt signaling in human and mouse breast cancer: Focusing on Wnt ligands, receptors and antagonists. Cancer Sci 2018;109:3368-75. [Crossref] [PubMed]
- Yang Q, Huang T, Ye G, et al. Methylation of SFRP2 gene as a promising noninvasive biomarker using feces in colorectal cancer diagnosis: a systematic meta-analysis. Sci Rep 2016;6:33339. [Crossref] [PubMed]
- Huyghe JR, Bien SA, Harrison TA, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet 2019;51:76-87. [Crossref] [PubMed]
- Zheng Y, Pan D. The Hippo Signaling Pathway in Development and Disease. Dev Cell 2019;50:264-82. [Crossref] [PubMed]
- Taniguchi K, Moroishi T, de Jong PR, et al. YAP-IL-6ST autoregulatory loop activated on APC loss controls colonic tumorigenesis. Proc Natl Acad Sci U S A 2017;114:1643-8. [Crossref] [PubMed]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017;36:1461-73. [Crossref] [PubMed]
- Ma D, Wei J, Chen S, et al. Fucoidan Inhibits the Progression of Hepatocellular Carcinoma via Causing lncRNA LINC00261 Overexpression. Front Oncol 2021;11:653902 [Crossref] [PubMed]
- Shi J, Ma H, Wang H, et al. Overexpression of LINC00261 inhibits non-small cell lung cancer cells progression by interacting with miR-522-3p and suppressing Wnt signaling. J Cell Biochem 2019;120:18378-87. [Crossref] [PubMed]
- Tang X, Sun Y, Wan G, et al. Knockdown of YAP inhibits growth in Hep-2 laryngeal cancer cells via epithelial-mesenchymal transition and the Wnt/β-catenin pathway. BMC Cancer 2019;19:654. [Crossref] [PubMed]
- Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene 2019;38:2627-44. [Crossref] [PubMed]
- Kim M, Han JH, Kim JH, et al. Secreted Frizzled-Related Protein 2 (sFRP2) Functions as a Melanogenic Stimulator; the Role of sFRP2 in UV-Induced Hyperpigmentary Disorders. J Invest Dermatol 2016;136:236-44. [Crossref] [PubMed]
- Huang T, Zhou Y, Zhang J, et al. The physiological role of Motin family and its dysregulation in tumorigenesis. J Transl Med 2018;16:98. [Crossref] [PubMed]
- Regan JL, Schumacher D, Staudte S, et al. Non-Canonical Hedgehog Signaling Is a Positive Regulator of the WNT Pathway and Is Required for the Survival of Colon Cancer Stem Cells. Cell Rep 2017;21:2813-28. [Crossref] [PubMed]
- Baharudin R, Tieng FYF, Lee LH, et al. Epigenetics of SFRP1: The Dual Roles in Human Cancers. Cancers (Basel) 2020;12:445. [Crossref] [PubMed]
- Tao X, Yang X, Wu K, et al. miR-629-5p promotes growth and metastasis of hepatocellular carcinoma by activating β-catenin. Exp Cell Res 2019;380:124-30. [Crossref] [PubMed]
- Heinosalo T, Gabriel M, Kallio L, et al. Secreted frizzled-related protein 2 (SFRP2) expression promotes lesion proliferation via canonical WNT signaling and indicates lesion borders in extraovarian endometriosis. Hum Reprod 2018;33:817-31. [Crossref] [PubMed]
- Jin Z, Chen B. LncRNA ZEB1-AS1 Regulates Colorectal Cancer Cells by MiR-205/YAP1 Axis. Open Med (Wars) 2020;15:175-84. [Crossref] [PubMed]
- Li H, Wang Z, Zhao G, et al. Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int J Biol Markers 2019;34:54-9. [Crossref] [PubMed]
- Liu X, Fu J, Bi H, et al. DNA methylation of SFRP1, SFRP2, and WIF1 and prognosis of postoperative colorectal cancer patients. BMC Cancer 2019;19:1212. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


