
Candidate oncogene circularNOP10 mediates gastric cancer progression by regulating miR-204/SIRT1 pathway
Introduction
The incidence of gastric cancer (GC) is declining worldwide (1-3). However, survival length and quality of life remain poor in some patients (1,4,5). Further, little is known about the pathogenesis and mechanism underlying the metastasis process. Investigation of the molecular events underlying the pathogenesis of GC is important for identifying novel therapeutic targets to improve GC survival rates.
Circular RNAs (circRNAs) are involved in several biological, pathological and physiological processes, such as cell cycle progression, cellular proliferation, as well as carcinogenesis and metastasis (6,7). Serving as microRNA (miRNA) sponges that prevented the target genes from miRNA-mediated mRNA dysfunction, circRNAs participate in the pathogenesis and progression of solid cancers (8,9) such as glioma (7), hepatocellular carcinoma (10), pancreatic carcinoma (11), GC (9,12), and colon cancer (13). The circRNAs have been reported to play biological roles in a variety of cancers, but little is known about its molecular mechanism and function in the pathogenesis of GC. Our previous study confirmed that miR-204 played a major role in regulating GC metastasis by affecting sirtuin 1 (SIRT1) expression (14). The present study used circRNA microarray analysis to clarify the relationship between circNOP10 (ID: hsa_circRNA-0034351, NM-018648) and miR-204 in regulating SIRT1 and GC progression.
SIRT1, a class III histone deacetylase (5,15,16), has been reported to be associated with the pathogenesis of neoplastic, metabolic, infectious, and neurodegenerative diseases (15,17). Recently, there has been some debate concerning the role of SIRT1 in the progression of GC (18). For example, some studies have found that SIRT1 expression was downregulated in GC patients with a poor prognosis (1,19). However, other studies have reported that SIRT1 levels in GC tissue were high, which is associated with epithelial-mesenchymal transition (EMT), cell invasion, as well as poor prognosis (5,12). Our previous paper supported the latter finding. However, details of the mechanisms in GC remain unclear (14). It is important that these conflicting functions of SIRT1 are better understood through studies of SIRT1 expression in cancer and its effects on well-known oncogenes and tumor suppressors (15,20). In the current study, we focused on the role of circNOP10 as an oncogene in GC progression. In addition, we determined the role of circNOP10 as a sponge for miR-204-mediated SIRT1 in the pathogenesis and progression of GC. Taken together, the present study revealed that circNOP10 showed oncogenic potency for the diagnosis and clinical management of GC. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-422).
Methods
Patients and samples
Tumorous tissues and paired adjacent nontumorous tissues were obtained from GC patients who underwent surgery in our hospital between February 2008 and February 2019. The tissue samples underwent immunohistochemistry (IHC) evaluation and Kaplan-Meier analysis were used to calculate survival estimates. In addition, SIRT1 and circNOP10 mRNA levels in the patients’ peripheral blood cells were measured. All patients signed a written informed consent. The study protocols were approved by the Ethical Committee of Zhongda Hospital, School of Medicine, Southeast University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell lines
Human epithelial gastric cell line GES-1 and GC cells including AGS, MNK-45, HGC-27 and BGC-823 were used in this study. Cells were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) and maintained in RPMI-1640 medium or F12 medium (Invitrogen, Waltham, MA, USA) supplemented with HyClone fetal bovine serum (10%, FBS). Cells were then incubated at 37 °C with 5% CO2 in a humid condition.
Microarray
Microarray was performed in 3 paired samples of GC fresh tissue and adjacent nontumorous tissue to analyze the expression profile of circRNA in GC. CircRNA microarray detection was performed by nKangchen Biotech (Shanghai, China).
Real-time PCR
Total RNA was extracted from cells, blood samples, and tissues using TRIzol reagent (Invitrogen Trading, Shanghai, China) according to the manufacturer’s instructions. Genomic DNA was extracted from cells using the PureLink Genomic DNA Mini Kit (Invitrogen Trading). Real-time PCR amplification for circNOP10 was conducted as previously described (14,21). Total RNA was extracted from the samples based on TRIzol (Invitrogen, 15596018), and then was treated with a wiper of genomic DNA. Afterwards, reverse transcription was conducted using the HiScript Q Select RT SuperMixKit (Vazyme Biotech, Nanjing, China), based on the stem-loop RT primer provided by Invitrogen. AceQ qPCR SYBR Green Master Mix (Vazyme) was utilized for the quantification of the products. Real-Time PCR was carried out to analyze the circNOP10 level that was normalized to U6. Mature circNOP10 and U6 primers were purchased from Invitrogen Trading. CircNOP10 underwent reverse transcription using the HiScript Q RT SuperMix for qPCR Kit (Vazyme) and quantified using SYBR Green Real-time PCR Master Mix. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the internal standard. The sequence for knockdown of circNOP10 expression was: human circNOP10 small interfering RNA(siRNA): 5'-GUCCAGAUAUUACAGGUAUTT-3'. The primer sequences for miR-204 and SIRT1 were shown in our previous study (14).
Western blot analysis
Western blot analysis was performed to assess the expression of SIRT1 [74465], E-cadherin [8426], Vimentin [6260], and β-actin (Abcam, ab6276), as previously described (14). All primary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).TGF-β was purchased from Sigma-Aldrich (St Louis, MO, USA).
RNA pulldown
The miR-204 probes were biotinylated, followed by transfecting into MNK-45 cells. About 48 h, the cells were harvested and lyzedon an icing condition. Dyna beads purchased from Thermo Fisher (Waltham, MA, USA) were incubated with the mixture of yeast transfer RNA and RNase-FBS to eliminate nonspecific binding. After incubating with the beads for 3 h at 4 °C, the samples were centrifuged at 3,000 rpm for 5 min to separate the beads. The buffer solution was utilized to wash the beads. Finally, the RNAs were extracted from the samples, and determined with the qRT-PCR according to the previous description (6).
Affinity isolation assay
The wash/binding buffer (500 mL) was utilized to dissolve the biotinylated DNA probes complementary to circNOP10. Then the DNA probes were incubated in M-280 Streptavid with magnetic beads at 4 °C for 3 h. The cell lysate of MNK-45 was incubated with the probe-coated beads for 1.5 h at 25 °C. Subsequently, the RNA complex bound to the beads was eluted and extracted. Upon washing with the wash/binding buffer, the RNA was subject to Real-time PCR analysis.
Transient transfection with synthetic miR-204-5p
MiR-204-5p inhibitor (anti-miR-204) together with the miR-204-5p inhibitor control (anti-miR-Con) were purchased from Ribobio (Guangzhou, China). MNK-45 or BGC-823 cells (1×105) were seeded in 24-well plates in 1 mL of antibiotic-free standard growth medium until a confluence of 60% was reached. Subsequently, the cells were transfected with miR-Con (50 nM), miR-204 (50 nM), anti-miR-Con (100 nM) or anti-miR-204 (100 nM). Then RiboFECT CP reagent (RiboBio, 1μL) was added into each well. About 24 h after transfection, the medium was replaced, and the transfection efficiency was determined through Western blot analysis.
Fluorescence in situ hybridization (FISH)
FISH was used to identify the localization of circNOP10 and miR-204 in MNK-45 cells. Hybridization was performed with circNOP10 and miR-204 probes. Ribo FISH Kit (Ribobio) was used for RNA FISH assay according to the manufacturer’s instructions. MNK-45 cells (1×104) were seeded onto coverslips in a 24-well plate, and washed with cold phosphate buffer saline (PBS) twice. The mixture was fixed with 4% paraformaldehyde for 20 min at room temperature and was then permeabilized using Triton X-100 (0.25%) for 15 min, followed by prehybridizing in hybridization solution at 37 °C for 1 h. Afterwards, labeled probes were added, and incubated with the cells in hybridization solution at 37 °C overnight. Blocking buffer was utilized for the incubation of coverslips, lasting for 1 h at room temperature. The mixture was incubated with primary antibodies including anti-digoxigenin-horseradish peroxidase or FITC at 4 °C overnight. Subsequently, the fluorescent secondary antibodies were added and mounted with 4',6-diamidino-2-phenylindole (DAPI) staining solution. The cell images were captured using a fluorescence microscope. The specificity of the circNOP10 and miR-204 signals were confirmed via comparison with a scrambled control.
Transwell assay
The invasion and migration of GC cells was carried out using Transwell assay. For the migration assay, the transfected cells (2×104) were transferred to the upper chamber, while the lower chamber was transferred with 10% FBS. About 48 h, invaded or migrated cells were fixed with methanol followed by staining with crystal violet (Sigma-Aldrich). Five fields were randomly selected to count the cells stained in color under a light microscope (Olympus, Tokyo, Japan). The procedures of cell invasion assay were the same way as the migration assay except the utilization of Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) in the upper chambers.
Cell proliferation, cell cycle and apoptosis assay
Cell proliferation was determined using Cell Counting Kit-8 (CCK-8, ab228554) at a wave-length of 450 nm. In terms of cell cycle analysis, PI/RNase staining buffer purchased from BD Bioscience was used to label the cells. Flow cytometry was conducted to measure the DNA content, with the aid of CellQuest software (BD Biosciences). For the analysis of cell apoptosis, cells were stained with propidium iodide (PI) (Sigma-Aldrich) and Annexin-V-FITC (BD Biosciences).
Statistical analysis
The association between SIRT1 expression and clinicopathological features of GC was evaluated using the Chi square test. The survival analysis was analyzed using the Kaplan-Meier plot and Log-rank test. The data were presented as mean ± standard deviation and were analyzed using Student’s t-test. Data analysis was performed using SPSS 21.0 software (IBM SPSS Statistics, Armonk, NY, USA). A value of P<0.05 was considered to be statistically significant.
Results
Upregulation of circNOP10 in GC tissues
Microarray was performed to analyze the expression profile of circRNA in GC based on 3 paired samples of GC tissues and para-carcinoma tissues. There were 60 differentially expressed circRNAs with a fold change of more than 1.5. The top 9 upregulated and 9 downregulated circRNAs were shown in Figure 1A. These 18 circRNAs were further identified in a cohort of GC tissues (n=10) using Real-time PCR, which indicated that hsa_circ-0034351 (circNOP10) was relatively higher among the 9 upregulated circRNAs (Figure 1B, Table 1) and the 9 downregulated circRNAs were higher in para-carcinoma than in cancer tissue (Figure 1C). Volcano plot and heat map showed these results in Figure 1D-1F. The hsa_circ-0034351 (circNOP10) level showed significant up-regulation (2.6-fold), we then investigated its roles in the pathogenesis of GC. We then explored the clinicopathologic significance of circNOP10 using RT-PCR analysis of 50 pairs of GC tissues and adjacent nontumorous tissues. As shown in Figure 1G, Kaplan-Meier survival curves revealed that GC patients with high expression of circNOP10 showed poorer overall survival rate (Table 2). Multi-variate analysis showed that circNOP10 level was an independent prognostic factor for GC (Figure 1H). These results demonstrated that the upregulation of circNOP10, a stable circRNA, was common in GC and responsible for poor prognosis. Human GC cell lines with high expression of circNOP10 (MNK-45 and BGC-823) were subsequently used for further investigation (Figure 1I). There was a high consistency between these data and the data obtained from microarray analysis, which indicated that the microarray data showed a high reliability.
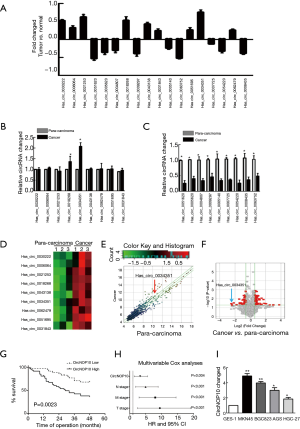

Full table

Full table
CircNOP10 knockdown downregulated SIRT1 expression
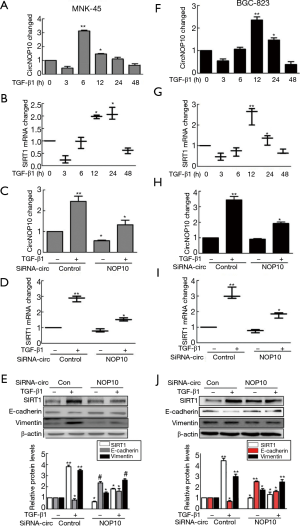
In order to explore the biological functions of circNOP10, transforming growth factor beta 1 (TGF-β1) was applied (24 hours, 5.0 ng/mL) (Figure S1A,B) and combined with siRNA to investigate the relationship between circNOP10 and SIRT1 in GC cell lines. As shown in Figure 2A,B, circNOP10 and SIRT1mRNA increased at different time intervals at transcriptional levels induced by TGF-β1. Moreover, circNOP10 and SIRT1mRNA were inhibited by circNOP10 siRNA at transcriptional levels by RT-PCR (Figure 2C,D). Compared to the control group, knockdown of circNOP10 resulted in downregulation of SIRT1 and Vimentin as well as upregulation of E-cadherin at post-transcriptional levels in MNK-45 cells (Figure 2E). Similar results were confirmed in BGC-823 cells (Figure 2F-2J).
CircNOP10 knockdown suppressed the malignant phenotype of GC cells
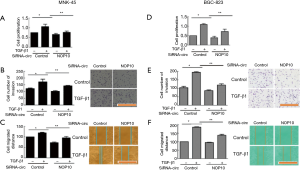
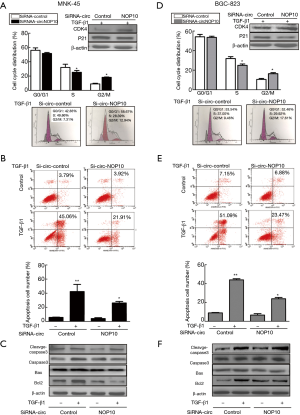
Given the fact that circNOP10 mediated EMT through SIRT1, we explored the biological function of circNOP10 in GC cell lines. The transduction of cells with circNOP10 siRNA significantly decreased cell proliferation and delayed the invasion of MNK-45 cells (Figure 3A-3C). Similar results further confirmed in BGC-823 cell lines (Figure 3D-3F). Additionally, in cell cycle distribution, knockdown of circNOP10 led to a decrease in the proportion of cells arrested in S-phase, which was accompanied by a concomitant increase of cell populations in the G2/M-phase (Figure 4A). Moreover, circNOP10 siRNA could significantly affect the stability of cyclin D1 protein and induce increased P21 expression in GC cells. Annexin-V-PI staining showed that circNOP10 affected the apoptosis of GC cell lines, reversed the number of apoptotic cells, upregulated the expression of apoptosis-related molecules, and activated the caspase-3 pathway in GC cells (Figure 4B and 4C). Similar effects were found in BGC-823 cells (Figure 4D-4F).These in vitro experiments suggested that circNOP10 might have played an oncogenic role in GC cell lines.
CircNOP10 acted as a molecular sponge for miR-204
Our previous data confirmed that the miR-204/SIRT1 axis mediated the pathogenesis and progression of GC. In the current study, we investigated whether circNOP10 could bind with miR-204. The RNAhybrid tool (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) confirmed that there was a miR-204 target site for circNOP10 (Figure 5A and 5B). We then explored the mechanism between circNOP10 and miR-204 in MNK-45 cells. In situ hybridization analysis using a circNOP10-specific probe showed, as expected, circNOP10 was expressed in MNK-45 cells (Figure 5C and 5D). MiR-204 and circNOP10 were colocalized in MNK-45 cells. RT-PCR was utilized to investigate whether circNOP10 involved in GC was induced by different stimuli. A biotin-coupled miR-204 mimic was used to test whether miR-204 was able to pull down circNOP10 expression (Figure 5E and 5F). We observed enrichment of circNOP10 in the miR-204 fractions following the introduction of mutations that disrupt base pairing between circNOP10 and miR-204. The circNOP10 and SIRT1mRNA showed increase in GC peripheral blood (Figure 5G and 5H). Taken together, these results demonstrated that circNOP10 was highly stable and was predominantly localized in the cytoplasm of MNK-45 cells.
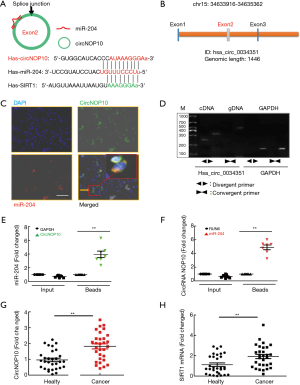
CircNOP10 sponged miR-204 mediating EMT in GC cells
In our previous study, negative feedback of miR-204 regulated SIRT1 expression and induced EMT in a clinical sample (Figure S2). We then hypothesized that SIRT1 might be a target of miR-204 in GC cells. Considering that circNOP10 can bind with miR-204, we then investigated the role of circNOP10 in EMT in GC cell lines. The transduction of cells with anti-miR-204 significantly ameliorated the upregulation of SIRT1 and Vimentin (Figure 6A). In contrast, increased E-cadherin was significantly reversed in TGF-β1-induced MNK-45 cells. Our previous results indicated that mimic-miR-204 triggered the downregulation of SIRT1 and upregulated E-cadherin at post-transcriptional levels (14). To verify whether miR-204 was a mediator of circNOP10, we co-transfected circNOP10 siRNA and anti-miR-204 in GC cells. As expected, co-transfection with circNOP10 siRNA and anti-miR-204 significantly decreased SIRT1 and Vimentin and enhanced E-cadherin levels in MNK-45 cells (Figure 6B). Moreover, the transduction of GC cells with circNOP10 siRNA resulted in downregulation of SIRT1 and Vimentin expression, which was inhibited by co-transduction with circNOP10 siRNA and anti-miR-204 in BGC-823 cells (Figure 6C and 6D). Taken together, circNOP10 could act as an endogenous miR-204 sponge involving in regulating the EMT via SIRT1 in GC cells.
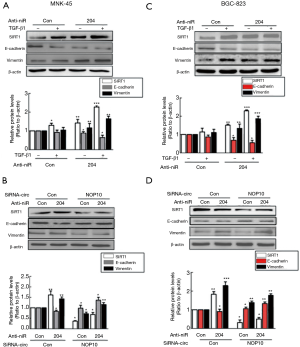
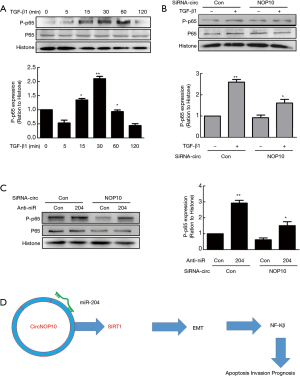
NF-κβ pathway involved in circNOP10-miR204-SIRT1 mediated EMT
Previous data reported that SIRT1 was involved in the NF-κβ signaling pathway in GC (18). Thus, subsequently, we examined whether this pathway was involved in EMT regulated by the circNOP10-miR204-SIRT1 axis. As shown in Figure 7A, TGF-β1 activated the NF-κβ level (phosphorylation of p65) in a time-dependent manner. Transduction of MNK-45 cells with circNOP10 siRNA significantly ameliorated the upregulation of phosphorylated p65 induced by TGF-β1 (Figure 7B). Moreover, the transduction of circNOP10 siRNA resulted in downregulation of NF-κβ, which was inhibited in cells co-transfected with circNOP10 siRNA and anti-miR204 (Figure 7C). Thus, we speculated that circNOP10 may serve as an endogenous miR-204 sponge for regulating the NF-κβ signaling pathway (Figure 7D).
Discussion
CircRNAs are increasingly receiving attention as rapid advances in high-throughput sequencing and bioinformatics technologies take place (22). There is accumulating evidence that circRNAs are involved in a variety of biological processes by acting as miRNA sponges or decoys, protein sponges or decoys, protein scaffolding, as well as an essential element in gene splicing and transcription (2,22,23). Moreover, circRNAs have been found to regulate tumor genesis by serving as translatable RNAs that are enriched in human cancers (4,17,24) upon binding with miRNAs (8). This might be related to the reversal of the inhibitory effects of miRNA downstream genes, which then participated in modulating the expression and function of target genes. For instance, circIFT80, as a ceRNA of miR-1236-3p, is reported to contribute to the progression of colorectal cancer (22). Upregulation of circRNA circ-000845 was associated with a poor prognosis among hepatocellular carcinoma patients as it could sponge miR-548 (25-27). In this study, there was an increase of novel circNOP10 in human GC tissues, indicating that circNOP10 could serve as a prognostic biomarker for GC. The knockdown of circNOP10 suppressed cell proliferation and invasion, the number of cells arrested in S-phase, cell apoptosis, and EMT. It is possible that circNOP10 acted as a competing endogenous RNA (ceRNA) by harboring miR-204, which attenuated the inhibitory effects of SIRT1 that promoted GC progression. On this basis, targeted circNOP10 blockade may serve as a potential therapeutic candidate for treating GC.
It has been widely acknowledged that circRNAs are involved in the pathogenesis and progression of several malignancies. CircRNAs have been identified in the conserved transcriptional groups of a large number of eukaryotic cells (28). Additionally, the expression of circRNAs is comparatively stable and is specifically expressed in certain tissues at a certain developmental stage (8,28). Importantly, it is relatively difficult for ribonuclease R (RNase R) to degrade circRNAs as they exhibit greater stability than linear RNAs in body fluids. Therefore, circRNAs show high potential value in clinical diagnosis and evaluation of prognosis (17). In this study, we evaluated the clinical relevance of circNOP10 in GC tissue. RT-qPCR revealed that elevation of circNOP10 was correlated with metastasis in lymphoid nodes among patients with advanced GC (Figure 1E). The level of circNOP10 in GC patients with poor prognosis was high, which was considered an independent prognostic factor for GC (Figure 1F). The combination of RT-qPCR and siRNA interference confirmed that high circNOP10 expression was involved in EMT by downregulating SIRT1 levels mediated by TGF-β1 (Figure 2E and 2G). These results indicated that circNOP10 localized at the upstream of SIRT1 could mediate EMT. In a recent study, circNOP10 regulated cell cycle progression through conjugation with CDKN1A/p21 and CDK2.Cell proliferative ability, Transwell assay and cell cycle distribution in GC cells are shown in Figure 2A-2D. In contrast, the decline of cell apoptosis and activation of the caspase-3 pathway induced by TGF-β1 were significantly reversed in MNK-45 cells transfected with circNOP10 siRNA. This indicated the potential role of circNOP10 as a marker in the diagnosis, treatment, and biological progression of GC.
CircRNAs, with the capacity to act as ceRNAs to sequester particular miRNAs family, could serve as competitive inhibitors to suppress its binding with the targets (20,29). For instance, circRNA CACTIN has been found to promote the GC progression by regulating TGFBR1 expression and binding with miR-331-3p (30). Furthermore, circRNA-0025202 was reported to modulate sensitivity to Tamoxifen and tumor progression by regulating the miR-182-5p/FOXO3a axis in patients with breast cancer (31). Our previous data confirmed that miR-204 negative feedback regulated SIRT1 expression in GC (14). We speculated that with a 2.6-fold change in GC tissue, circNOP10 might function as a ceRNA. The combination of multiple bioinformatic analyses and RNA pulldown skills confirmed miR-204 as a tumor-suppressing miRNA in GC with potential binding sites for circNOP10. FISH assay was carried out to verify whether miR-204 was a mediator of circNOP10, with the results indicating that the transduction of MNK-45 with anti-miR-204 significantly increased the SIRT1 and E-cadherin expression induced by TGF-β1. As expected, the transduction of MNK-45 with mimic-miR204 significantly inhibited the upregulation of SIRT1 and Vimentin. Moreover, such phenomenon was reversed in co-transfected cells, with circNOP10 siRNA and anti-miR204. This indicated that circNOP10 acted as a miR-204 sponge to regulate SIRT1 expression and attenuate EMT in GC cells. Our data indicated that circNOP10 could directly bind to miR-204 and might have served as a ceRNA to mediate EMT viaSIRT1. More studies are required to clarify the mechanism and function of circNOP10.
SIRT1 has a dual role by functioning as a tumor suppressor gene and an oncogene (25). To date, there is still debate over the expression of SIRT1 in different cancers. The expression of SIRT1 was relatively high in patients with breast cancer, hepatocellular carcinoma, and thyroid cancer (27,30,31), but was lower in those with colon cancer and lung cancer (32,33) when comparing with the adjacent tissues. Even in the same type of cancer (for example, prostate cancer), the results are controversial (25,26). On this basis, we assumed that the roles of SIRT1 in the oncogenic progression were highly depending on the type of malignancies. In this study, we detected the expression of SIRT1 in GC, which showed that both the mRNA and the protein of SIRT1 showed significant increase compared with the matched normal tissue. Furthermore, SIRT1 also participated in invasion, proliferation, and EMT in GC cells (1,32,34). Western blot assays demonstrated elevation of SIRT1 and E-cadherin expression, which then triggered transcriptional activation and alternations of cell cycle distribution, as well as cell apoptosis and downregulation of cell proliferation (35,36). Interestingly, these findings were not consistent with the results of a previous study (28). Although the same antibody for SIRT1 was used, there might be differences among the samples obtained from patients of different races.
SIRT1 can inhibit the activity of E2F1, a cell-cycle and apoptosis regulator (37), which would then induce cell proliferation and cell-arrest in the G1 phase by inhibiting cyclin D1 (38), following NF-κβ/Cyclin D1 pathway inhibition (18) in cancer (39,40). Our results indicated that TGF-β1 activated the NF-κβ pathway and increased the p-p65/p65 ratio in MNK-45 cells. In addition, circNOP10 knockdown significantly attenuated p-p65 levels. The combination of siRNA circNOP10 and miR-204 inhibitor significantly attenuated p-p65 levels on the NF-κβ pathway. These results demonstrated that circNOP10/miR-204/SIRT1 promoted GC progression via the NF-κβ pathway in GC cells. For the first time, our study validated that the circNOP10/miR204/SIRT1 axis might control the progression of GC and that circNOP10 may be a promising therapeutic target for the treatment of GC.
Conclusions
This study found that CircNOP10 promoted GC cell proliferation, invasion, apoptosis, EMT, and prognosis. The ceRNA regulatory network of circNOP10 could target miRNA-204 and SIRT1, which indicated that they could serve as biomarkers for the diagnosis and targeted therapy among GC patients.
Acknowledgments
Funding: This research was supported by the Jiangsu Provincial Natural Science Fund (Grant No.: BK2012750) and the Fundamental Research Funds for the Central Universities and the College Graduate Research and Innovation program of Jiangsu Province (Grant No.: KYLX15_0179).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-422
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-422
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-422). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients signed a written informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocols were approved by the Ethical Committee of Zhongda Hospital, School of Medicine, Southeast University (No: 2019ZDSYLL067-P01). All patients signed a written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- An Y, Wang B, Wang X, et al. SIRT1 inhibits chemoresistance and cancer stemness of gastric cancer by initiating an AMPK/FOXO3 positive feedback loop. Cell Death Dis 2020;11:115. [Crossref] [PubMed]
- Cai X, Nie J, Chen L, et al. Circ_0000267 promotes gastric cancer progression via sponging MiR-503-5p and regulating HMGA2 expression. Mol Genet Genomic Med 2020;8:e1093 [Crossref] [PubMed]
- Baj J, Brzozowska K, Forma A, et al. Immunological Aspects of the Tumor Microenvironment and Epithelial-Mesenchymal Transition in Gastric Carcinogenesis. Int J Mol Sci 2020;21:2544. [Crossref] [PubMed]
- Jiang Y, Zhang Y, Chu F, et al. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int 2020;20:74. [Crossref] [PubMed]
- Özcan Ö, Belli AK, Sakallı Çetin E, et al. Upregulation of SIRT1 gene in gastric adenocarcinoma. Turk J Gastroenterol 2019;30:326-30. [Crossref] [PubMed]
- Hu K, Qin X, Shao Y, et al. Circular RNA MTO1 suppresses tumorigenesis of gastric carcinoma by sponging miR-3200-5p and targeting PEBP1. Mol Cell Probes 2020;52:101562 [Crossref] [PubMed]
- Lei B, Huang Y, Zhou Z, et al. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression. J Cell Biochem 2019;120:6698-708. [Crossref] [PubMed]
- Deng G, Mou T, He J, et al. Circular RNA circRHOBTB3 acts as a sponge for miR-654-3p inhibiting gastric cancer growth. J Exp Clin Cancer Res 2020;39:1. [Crossref] [PubMed]
- Quan J, Dong D, Lun Y, et al. Circular RNA circHIAT1 inhibits proliferation and epithelial-mesenchymal transition of gastric cancer cell lines through downregulation of miR-21. J Biochem Mol Toxicol 2020;34:e22458 [Crossref] [PubMed]
- Fu X, Zhang J, He X, et al. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J Cancer 2020;11:3318-26. [Crossref] [PubMed]
- Chen G, Shi Y, Zhang Y, et al. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther 2017;10:5783-94. [Crossref] [PubMed]
- Li H, He C, Wang X, et al. MicroRNA-183 affects the development of gastric cancer by regulating autophagy via MALAT1-miR-183-SIRT1 axis and PI3K/AKT/mTOR signals. Artif Cells Nanomed Biotechnol 2019;47:3163-71. [Crossref] [PubMed]
- He JH, Han ZP, Luo JG, et al. Hsa_Circ_0007843 Acts as a mIR-518c-5p Sponge to Regulate the Migration and Invasion of Colon Cancer SW480 Cells. Front Genet 2020;11:9. [Crossref] [PubMed]
- Zhang L, Wang X, Chen P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer 2013;13:290. [Crossref] [PubMed]
- Noguchi A, Kikuchi K, Zheng H, et al. SIRT1 expression is associated with a poor prognosis, whereas DBC1 is associated with favorable outcomes in gastric cancer. Cancer Med 2014;3:1553-61. [Crossref] [PubMed]
- Ota H, Tokunaga E, Chang K, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene 2006;25:176-85. [Crossref] [PubMed]
- Wu J, Chen Z, Song Y, et al. CircRNA_0005075 suppresses carcinogenesis via regulating miR-431/p53/epithelial-mesenchymal transition axis in gastric cancer. Cell Biochem Funct 2020;38:932-42. [Crossref] [PubMed]
- Yang Q, Wang B, Gao W, et al. SIRT1 is downregulated in gastric cancer and leads to G1-phase arrest via NF-κB/Cyclin D1 signaling. Mol Cancer Res 2013;11:1497-507. [Crossref] [PubMed]
- Mohammadi Saravle S, Ahmadi Hedayati M, Mohammadi E, et al. Sirt1 Gene Expression and Gastric Epithelial Cells Tumor Stage in Patients with Helicobacter pylori Infection. Asian Pac J Cancer Prev 2018;19:913-6. [PubMed]
- Xu G, Chen Y, Fu M, et al. Circular RNA CCDC66 promotes gastric cancer progression by regulating c-Myc and TGF-β signaling pathways. J Cancer 2020;11:2759-68. [Crossref] [PubMed]
- Zhang Y, Shen K, Bai Y, et al. Mir143-BBC3 cascade reduces microglial survival via interplay between apoptosis and autophagy: Implications for methamphetamine-mediated neurotoxicity. Autophagy 2016;12:1538-59. [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Lu Y, Tan L, Wang X. Circular HDAC9/microRNA-138/Sirtuin-1 Pathway Mediates Synaptic and Amyloid Precursor Protein Processing Deficits in Alzheimer's Disease. Neurosci Bull 2019;35:877-88. [Crossref] [PubMed]
- Hua X, Sun Y, Chen J, et al. Circular RNAs in drug resistant tumors. Biomed Pharmacother 2019;118:109233 [Crossref] [PubMed]
- Bosch-Presegué L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer 2011;2:648-62. [Crossref] [PubMed]
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci 2009;5:147-52. [Crossref] [PubMed]
- Eades G, Yao Y, Yang M, et al. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem 2011;286:25992-6002. [Crossref] [PubMed]
- Shi JQ, Wang B, Cao XQ, et al. Circular RNA_LARP4 inhibits the progression of non-small-cell lung cancer by regulating the expression of SMAD7. Eur Rev Med Pharmacol Sci 2020;24:1863-9. [PubMed]
- Yao Y, Hua Q, Zhou Y, et al. CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin signaling pathway. Biomed Pharmacother 2019;111:1367-75. [Crossref] [PubMed]
- Herranz D, Maraver A, Cañamero M, et al. SIRT1 promotes thyroid carcinogenesis driven by PTEN deficiency. Oncogene 2013;32:4052-6. [Crossref] [PubMed]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature 2008;451:583-6. [Crossref] [PubMed]
- Kabra N, Li Z, Chen L, et al. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem 2009;284:18210-7. [Crossref] [PubMed]
- Beane J, Cheng L, Soldi R, et al. SIRT1 pathway dysregulation in the smoke-exposed airway epithelium and lung tumor tissue. Cancer Res 2012;72:5702-11. [Crossref] [PubMed]
- Byles V, Zhu L, Lovaas JD, et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene 2012;31:4619-29. [Crossref] [PubMed]
- Pruitt K, Zinn RL, Ohm JE, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet 2006;2:e40 [Crossref] [PubMed]
- Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res 2009;69:1702-5. [Crossref] [PubMed]
- Wang C, Chen L, Hou X, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol 2006;8:1025-31. [Crossref] [PubMed]
- Mao B, Zhao G, Lv X, et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol 2011;43:1573-81. [Crossref] [PubMed]
- Chan KS, Koh CG, Li HY. Mitosis-targeted anti-cancer therapies: where they stand. Cell Death Dis 2012;3:e411 [Crossref] [PubMed]
- Starostina NG, Kipreos ET. Multiple degradation pathways regulate versatile CIP/KIP CDK inhibitors. Trends Cell Biol 2012;22:33-41. [Crossref] [PubMed]
(English Language Editor: A. Muijlwijk)


