Risk characteristics with seven epithelial-mesenchymal transition-related genes are used to predict the prognosis of patients with hepatocellular carcinoma
Introduction
As the 6th most common cancer worldwide, first-time liver cancer is one of the most common malignant tumors (1). It is also the 4th leading cause of cancer death. The disease is mainly caused by hepatitis B virus (HBV) or hepatitis C virus, infection, alcoholism, obesity, and nonalcoholic fatty liver disease. With an incidence of 90%, liver cancer is the most common pathological hepatocellular carcinoma (HCC). In the early stage of liver cancer, patients may be unaware of their diagnoses, as there are no obvious symptoms. Its development and transfer are very rapid. At the time of diagnosis, most patients are locally advanced or have distant metastases. Most advanced patients cannot undergo surgery, and the recurrence rate of early patients after 5 years is 50% (2). In general, the prognosis of HCC patients is poor. Even with an accepted diagnosis of HCC and rapid treatment, it is difficult to extend survival time because of its high metastasis rate and recurrence rate. Numerous researchers have sought to find a high-precision diagnostic method to improve the detection rate of early liver cancer and extend the survival time and quality of life of patients with liver cancer (3).
The metastasis of cancer cells is key to the high recurrence and mortality of HCC; however, the mechanism by which metastasis occurs is not clear (4). The activation of epithelial-mesenchymal transition (EMT) is key to metastasis of tumor cells; 90% of cells in human tumors are epithelial . Under this EMT process, epithelial cells acquire the characteristics of mesenchymal cells, and the ability of cell movement and migration is enhanced (5). Pathologically, tumor cells grow in the primary site, invade surrounding normal tissues, penetrate the basement membrane, invade blood vessels, enter the circulatory system, survive to reach remote sites, invade blood vessels, and form new metastatic sites.
Databases containing the gene expression data and follow-up data of patients have been established (6). At present, most bioinformatic researchers are trying to develop bioinformatic analysis methods using the corresponding genome database to screen EMT-related genes (ERGS) with a predictive index value for prognosis, and establish a variety of prognostic models that can predict cancer. However, the correlation between ERGS and HCC prediction has not been studied (7).
HCC patients with similar clinicopathological features have different prenatal and postpartum symptoms. Thus, the possible mechanism by which the spread of liver cancer can be inhibited needs to be identified, and a new mechanism related to the precursor symptoms of Adefovir (ADV) found. Meanwhile, we recognize the treatment of patients with HCC and the postpartum assessment of the high incidence and mortality associated with HCC is essential. According to relevant reports in the literature, the early spread of HCC is a key factor for recurrence and death from HCC patients (8). However, despite numerous studies, the mechanism underlying the early metastasis of HCC is still very difficult to understand.
To control the transfer of HCC, EMT should be used. In previous studies, we confirmed the presence of 38-RNF in the ring finger . In HCC, the induction of EMT could potentially promote tumor invasion and metastasis. By promoting transforming growth factor-beta, RNF38 could induce HCC cells EMT, thereby promoting the invasion and metastasis of HCC cells in the body. During EMT, epidermal cells may lose contact with each other due to the melting of E-cadherin, which in turn increases the possibility of migration, invasion, and cancer transmission in direct and indirect tissues (9). This is an important early stage of HCC. This study showed that a large number of genes associated with EMT can predict HCC.
With the emergence of the micro-matrix and sequence method, the scientific community has constructed genomic databases for multiple patients (10). Using these databases, many studies have been carried out on malignant and normal tissues to determine the predictability of ERGS, and establish prognostic models that use relevant genomes to predict a variety of cancers. However, the preliminary value of ERGS in HCC has not been fully studied. In this study, we used data from The Cancer Genome Atlas (TCGA) database, identified genes and examined the clinical prognostic effects of HCC . Prognostic predictors were derived from patients identified to have prognostically relevant ERGS (11). We used these data to establish the expression levels of ERGS and formulae to generate a risk score for each HCC patient and predict and assess the prognosis of each HCC patient (12).
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-394).
Methods
Data acquisition
Epithelial-mesenchymal transition (EMT) gene were obtained from dbEMT2.0 database (http://dbemt.bioinfo-minzhao.org/download.cgi). Totally, we obtained 1,184 EMT-related genes (ERGs) from the dbEMT2.0 database. RNA-seq data, as also the clinical information for LIHC were obtained from the TCGA database (https://portal.gdc.cancer.gov/). Similarly, The LIRI-JP dataset was obtained from the ICGC (https://daco.icgc.org/) for the validation studies (13). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differentially expressed ERGs and enrichment analysis
We use the edgeR package in the R software to analyze hepatocellular tumor samples and normal tissue samples to identify differentially expressed ERGs, with adjusted P<0.05 and |log2FC| >1. Volcano plots and heat maps were visualized base on the ggplot2 and pheatmap packages in R software. Evaluate the potential molecular mechanism of ERGS in cancer through GO analysis, KEGG analysis and GSEA analysis. The results are visualized using the clusterProfiler and richplot packages in R.
Construction and validation of ERG signature
In order to identify genes related to OS with P values <0.01, Univariate Cox regression analysis was used. Then, the significant prognostic genes were filtered by multivariate Cox regression analysis. Following the results of multivariate Cox regression analyses, a prognosis-predictive formula was generated: Risk score = expression of gene 1× G1 + expression of gene 2× G2 + expression of gene n× Gn, where Gn represents the prognostic gene. Then We used Kaplan-Meier survival curves and the logrank method to estimate the prognostic significance. A P value <0.05 was considered statistically significant. Receiver operating characteristic (ROC) analyses were performed in R survival ROC to validate gene signatures. Subsequently, the patients with LIHC were classified into low- and high-risk groups in either cohort based on the median risk
Development of nomogram
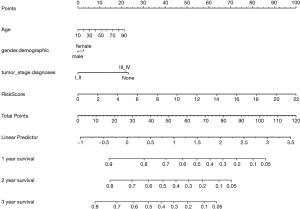
Age, gender, stage, and risk score feature in this cohort were used to construct a nomogram, using the survival and rms package for R. Moreover, calibration curves were plotted to assess the concordance between actual and predicted survival (14).
Statistical analysis
All statistical analyses were applied with R (version 3.6.0). The mRNA expression level was log2-normalized for analysis. Kaplan-Meier analysis with a logrank test (15) was performed for comparison between the low- and high-risk groups. Univariate and multivariate Cox proportional hazard regression analyses were performed to determine prognostic values for risk scores, as well as various clinical features. P value <0.05 was considered to indicate statistical significance (16).
Results
Identification of differentially expressed ERGs
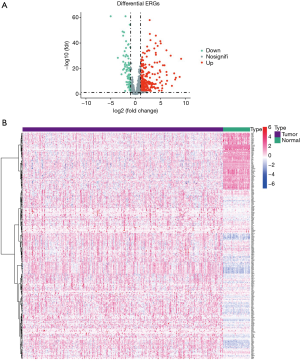
First, we downloaded data and clinical information about HCC patients from TCGA database, and extracted genes from 400 HCC samples and 60 common samples. To obtain the clinicopathological data of HCC patients, including age, gender, organizational education, vocational training, and tumor, node, and metastasis (TNM) stage (17) data, we screened 393 HCC patients from the 400 HCC samples. A set of genes from the dbEMT2.0 with the marker for EMTs was then selected, showed in https://cdn.amegroups.cn/static/public/jgo-21-394-1.xlsx. We then used the software package to detect and analyze the positive expression of RNA and check the Lima Wilcoxon symbol level (FDR <0.05, log2FC >1). We then selected the differentially expressed genes (DEGs) between HCC and normal samples. From these results, we screened 12 downregulated ERGs and 84 upregulated ERGs from the liver cancer samples (see Figure 1). The expression levels of these DEGs in HCC and normal liver tissues were showed by a volcano plot and heatmap plot (18,19).
Biological functions and significant pathway analysis
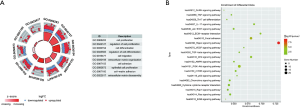
After the GO enrichment and KEGG analyses, the molecular mechanism and biological effects of the 96 ERGs were studied. According to the results of the GO analysis, ERGs are mainly involved in biological processes related to tissue or embryo development (see Figure 2A). In addition, the KEGG analysis showed that these 96 ERGs interacted with extracellular matrix (ECM) receptors and were associated with cancer proteoglycans and oncogenic phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway (Figure 2B).
Screening hub ERGs related to prediction model
After screening for differences in ERGs, Univariate Cox analysis and multiple Cox regression analysis methods were used to verified whether the model could be independent progress factors.
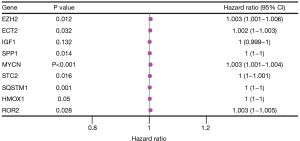
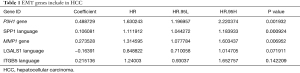
Univariate Cox model results 32 ERGS were significantly. Then a multivariate regression analysis was conducted to determine and obtain the regression coefficients related to the model. Results showed 7 prognostically relevant ERGs [i.e., ECT2, EZH2, MYCN, ROR2, secretory phosphoprotein 1 (SPP1), SQSTM1, and STC2] were significantly (see Figure 3).
Establishment and validation of the prognostic model
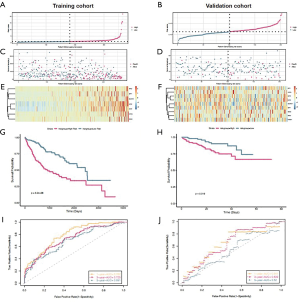
After Univariate Cox analysis and multiple Cox regression analysis, we obtained a list of differentially expressed ERGS and then used the selected ERGS and their regression coefficients to develop a prognostic prediction model for HCC patients (Table 1). The following formula was applied to reveal the ERGS-related prognostic risk model: (0.488729 × P3H1) + (0.106081 × SPP1) + (0.273528 × MMP1) – (0.16391 × LGALS1) + [0.215136 × integrin-β5 (ITGB5)]. According to the median risk score (20-23), the samples were divided into high-risk groups and low-risk groups. The results show that in the training and validation cohorts, higher risk scores are correlated with the quality of life of HCC patients (Figure 4A-4F). The Kaplan-Meier analysis displayed a significant difference in the outcome of the patients between the high-risk group and the low-risk group (log-rank test P<0.001; Figure 4G,4H). Next, receiver operating characteristic (ROC) curve analysis was carried out to assess the discrimination capacity in training and validation cohort. The areas under the curves (AUCs) for 1-, 3-, and 5-year OS predictions in training cohort and validation cohort were 0.782, 0.725, 0.691 and 0.641, 0.639, 0.52, respectively (Figure 4I,4J), which indicated a good performance of these signature for prognostic prediction (24,25).
Full table
Construction of a nomogram
We have developed a schematic diagram based on risk models for the clinical design and application (26-29). We analyzed and calculated the overall scores used to predict the 1-, 2-, and 3-year overall survival of HCC patients (30). According to Cox regression combined with critical clinical parameters, the nomogram contains three prognostic parameters: age, gender, stage, and risk score (Figure 5). Each patient gets the point for each prognostic parameter, and the higher the total score, the worse the result.
Subgroup analysis of the ERG signature
In order to find difference between to subtype. The prognostic value of the ERG signature was used to explored in subgroups of patients with HCC with various clinical and demographic features. Stratification analysis was carried out according to the clinical features, including gender, pathology metastasis, stage (Figure 6).

Discussion
HCC is a malignant tumor with a high recurrence rate and poor prognosis (31). Previously, the clinicopathological features of HCC patients have been used to predict their prognosis (32); however, these features do not accurately assess prognosis. Later, serum tumor markers were used for prediction; however, their accuracy is similarly insufficient. Metastasis is a common feature of many cancers and an important factor leading to poor prognosis and death in patients with liver cancer. Metastasis is associated with poor outcomes and mortality in patients with HCC. ERGs have the potential to predict the prognosis of some tumors. Thus, we sought to use ERGs to construct a prognosis prediction model for liver cancer patients.
Research has shown that ERGs have good reliability in predicting potential. Thus, it is necessary to research and explore an advanced strategy based on prognosis assessment in HCC patients with ERGs. We constructed a prognosis prediction model based on TCGA database (33) and clarified the prognostic value of global ERGs. First, 1,184 EAG lists from dbEMT2 database were selected based on information in TCGA database. From the ERGs lists, 12 downregulated and 84 upregulated ERGs were selected. GO enrichment and KEGG analyses were performed to reveal the underlying biological pathways and processes associated with these ERGs (34-36).
By analyzing the gene sequencing data and clinical prognosis information of HCC patients, a predictive risk model was constructed based on the five prognostic ERGS (i.e., LGALS1, MMP1, P3H1, ITGB5, and SPP1). The results of univariate and multivariate regression analysis show that our risk model can be used as an independent prognostic model, can predict the prognosis well, and accurately assess the overall survival rate of HCC patients (37,38). The predictive value of this model was verified using Kaplan-Meier and ROC curves. The Kaplan-Meier curves showed that the overall survival rate of patients in the low-risk group was significantly higher than that of patients in the high-risk group (P<0.00001). In addition, compared to other clinical characteristics, the area under the curve (AUC) value of the risk model was significantly higher (AUC =0.723). Compared to other clinical features, this model is more accurate at predicting the prognosis of HCC patients. Finally, a histogram was constructed to generate a model a risk score, the clinical characteristics of patients were constructed, and overall survival was directly predicted (39).
Previous studies have shown that the development and prognosis of patients with HCC are associated with some of these 5 EAG hubs. For example, one member of the integrin family, ITGB5, promotes tumorigenicity in HCC. Thus, potential independent prognostic biomarkers may exist in HBV-associated HCC patients. In addition, gene metalloproteinase 1 (MMP1), a member of zinc-dependent endocletic proteinases, degrades the ECM. These genes regulate the EMT process in HCC by disrupting the structural proteins. Poor prognosis in HCC has also been associated with the high expression of MMP1 (40). In addition, SPP1 integrin binding is involved in overexpression in many cancers, including HCC. Results showed that the prognostic value of HCC, SPP1 has been widely studied and verified. However, LGALS1, also known as galectin-1, is highly expressed in some tumors (e.g., glioma, leukemia, and oral cancer), and regulates the immune response, cancer metastasis, and cell viability. So far, the role of LGALS1 in HCC has not been studied to a large extent. Similarly, no evidence was found for the role of P3H1 in HCC. Through bioinformatics analysis, we discovered the role of LGALS1 and P3H1 in predicting HCC and its prognostic value, which may help other researchers to study HCC and provide new ideas and discover new biomarkers for the prognosis of HCC patients (41).
Sequencing data of ERGs and clinical information from TCGA were used to construct a reliable database of prognostic risk models for HCC patients. Compared to other clinicopathological features, we achieved satisfactory results in terms of the prognostic predictive function of the risk model. Additionally, we used Kaplan-Meier and ROC curves for further investigations. Notably, this may be the leading study in the prognosis of HCC patients and the expression of their ERGs. It provides a new method to predict and assess liver cancer prognosis with high accuracy and precision (42).
Conclusions
In short, we built a risk prediction model using 7 ERGS and nomograms . This new method detects the expression of ERGS to predict patient clinical information. Compared with traditional methods, this risk model method can achieve better prognosis prediction and supervision for HCC patients. Our findings provide new insights into the relationship between hepatocellular carcinoma development and EMT.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-394
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-394). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen Q, Han B, Meng X, et al. Immunogenomic analysis reveals LGALS1 contributes to the immune heterogeneity and immunosuppression in glioma. Int J Cancer 2019;145:517-30. [Crossref] [PubMed]
- Jo H, Lee J, Jeon J, et al. The critical role of glucose deprivation in epithelial-mesenchymal transition in hepatocellular carcinoma under hypoxia. Sci Rep 2020;10:1538. [Crossref] [PubMed]
- Kim J, Hong SJ, Park JY, et al. Epithelial-mesenchymal transition gene signature to predict clinical outcome of hepatocellular carcinoma. Cancer Sci 2010;101:1521-8. [Crossref] [PubMed]
- Chen S, Zhao E. Development and validation of a robust epithelial-mesenchymal transition (EMT)-related prognostic signature for hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2021;45:101587 [Crossref] [PubMed]
- Kong F, Zhou K, Zhu T, et al. Interleukin-34 mediated by hepatitis B virus X protein via CCAAT/enhancer-binding protein α contributes to the proliferation and migration of hepatoma cells. Cell Prolif 2019;52:e12703 [Crossref] [PubMed]
- Lee JH, Jung S, Park WS, et al. Prognostic nomogram of hypoxia-related genes predicting overall survival of colorectal cancer-Analysis of TCGA database. Sci Rep 2019;9:1803. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Liao M, Tong P, Zhao J, et al. Prognostic value of matrix metalloproteinase-1/ proteinase-activated receptor-1 signaling axis in hepatocellular carcinoma. Pathol Oncol Res 2012;18:397-403. [Crossref] [PubMed]
- Li JM, Tseng CW, Lin CC, et al. Upregulation of LGALS1 is associated with oral cancer metastasis. Ther Adv Med Oncol 2018;10:1758835918794622 [Crossref] [PubMed]
- Fu XT, Dai Z, Song K, et al. Macrophage-secreted IL-8 induces epithelial-mesenchymal transition in hepatocellular carcinoma cells by activating the JAK2/STAT3/Snail pathway. Int J Oncol 2015;46:587-96. [Crossref] [PubMed]
- Tao YM, Huang JL, Zeng S, et al. BTB/POZ domain-containing protein 7: epithelial-mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology 2013;57:2326-37. [Crossref] [PubMed]
- Lin Z, He R, Luo H, et al. Integrin-β5, a miR-185-targeted gene, promotes hepatocellular carcinoma tumorigenesis by regulating β-catenin stability. J Exp Clin Cancer Res 2018;37:17. [Crossref] [PubMed]
- Long J, Zhang L, Wan X, et al. A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma. J Cell Mol Med 2018;22:5928-38. [Crossref] [PubMed]
- Hu B, Cheng JW, Hu JW, et al. KPNA3 Confers Sorafenib Resistance to Advanced Hepatocellular Carcinoma via TWIST Regulated Epithelial-Mesenchymal Transition. J Cancer 2019;10:3914-25. [Crossref] [PubMed]
- Tong H, Liu X, Li T, et al. NR1D2 Accelerates Hepatocellular Carcinoma Progression by Driving the Epithelial-to-Mesenchymal Transition. Onco Targets Ther 2020;13:3931-42. [Crossref] [PubMed]
- Liu L, Dai Y, Chen J, et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial-mesenchymal transition by way of Akt/GSK-3β/Snail signaling. Hepatology 2014;59:531-43. [Crossref] [PubMed]
- Ma P, Tang WG, Hu JW, et al. HSP4 triggers epithelial-mesenchymal transition and promotes motility capacities of hepatocellular carcinoma cells via activating AKT. Liver Int 2020;40:1211-23. [Crossref] [PubMed]
- Ouyang G, Yi B, Pan G, et al. A robust twelve-gene signature for prognosis prediction of hepatocellular carcinoma. Cancer Cell Int 2020;20:207. [Crossref] [PubMed]
- Tu Y, Chen C, Fan G. Association between the expression of secreted phosphoprotein - related genes and prognosis of human cancer. BMC Cancer 2019;19:1230. [Crossref] [PubMed]
- Ruvolo PP. Galectins as regulators of cell survival in the leukemia niche. Adv Biol Regul 2019;71:41-54. [Crossref] [PubMed]
- Umezaki N, Nakagawa S, Yamashita YI, et al. Lysyl oxidase induces epithelial-mesenchymal transition and predicts intrahepatic metastasis of hepatocellular carcinoma. Cancer Sci 2019;110:2033-43. [Crossref] [PubMed]
- Huang XY, Ke AW, Shi GM, et al. αB-crystallin complexes with 14-3-3ζ to induce epithelial-mesenchymal transition and resistance to sorafenib in hepatocellular carcinoma. Hepatology 2013;57:2235-47. [Crossref] [PubMed]
- Scheau C, Badarau IA, Costache R, et al. The Role of Matrix Metalloproteinases in the Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma. Anal Cell Pathol (Amst) 2019;2019:9423907 [Crossref] [PubMed]
- van Zijl F, Zulehner G, Petz M, et al. Epithelial-mesenchymal transition in hepatocellular carcinoma. Future Oncol 2009;5:1169-79. [Crossref] [PubMed]
- Shang L, Ye X, Zhu G, et al. Prognostic value of integrin variants and expression in post-operative patients with HBV-related hepatocellular carcinoma. Oncotarget 2017;8:76816-31. [Crossref] [PubMed]
- Wang H, Xu H, Ma F, et al. Zinc finger protein 703 induces EMT and sorafenib resistance in hepatocellular carcinoma by transactivating CLDN4 expression. Cell Death Dis 2020;11:225. [Crossref] [PubMed]
- Altadill A, Rodríguez M, González LO, et al. Liver expression of matrix metalloproteases and their inhibitors in hepatocellular carcinoma. Dig Liver Dis 2009;41:740-8. [Crossref] [PubMed]
- Chen PF, Li QH, Zeng LR, et al. A 4-gene prognostic signature predicting survival in hepatocellular carcinoma. J Cell Biochem 2019;120:9117-24. [Crossref] [PubMed]
- Zhang XP, Jiang YB, Zhong CQ, et al. PRMT1 Promoted HCC Growth and Metastasis In Vitro and In Vivo via Activating the STAT3 Signalling Pathway. Cell Physiol Biochem 2018;47:1643-54. [Crossref] [PubMed]
- Yang Z, Qi Y, Lai N, et al. Notch1 signaling in melanoma cells promoted tumor-induced immunosuppression via upregulation of TGF-β1. J Exp Clin Cancer Res 2018;37:1. [Crossref] [PubMed]
- Yin X, Zhang BH, Zheng SS, et al. Coexpression of gene Oct4 and Nanog initiates stem cell characteristics in hepatocellular carcinoma and promotes epithelial-mesenchymal transition through activation of Stat3/Snail signaling. J Hematol Oncol 2015;8:23. [Crossref] [PubMed]
- Wang J, Hao F, Fei X, et al. SPP1 functions as an enhancer of cell growth in hepatocellular carcinoma targeted by miR-181c. Am J Transl Res 2019;11:6924-37. [PubMed]
- Wang TH, Lin YS, Chen Y, et al. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget 2015;6:23342-57. [Crossref] [PubMed]
- Wang L, Sun L, Wang Y, et al. miR-1204 promotes hepatocellular carcinoma progression through activating MAPK and c-Jun/AP1 signaling by targeting ZNF418. Int J Biol Sci 2019;15:1514-22. [Crossref] [PubMed]
- Wang X, Yao S, Xiao Z, et al. Development and validation of a survival model for lung adenocarcinoma based on autophagy-associated genes. J Transl Med 2020;18:149. [Crossref] [PubMed]
- Zhang T, Guo J, Gu J, et al. KIAA0101 is a novel transcriptional target of FoxM1 and is involved in the regulation of hepatocellular carcinoma microvascular invasion by regulating epithelial-mesenchymal transition. J Cancer 2019;10:3501-16. [Crossref] [PubMed]
- Xia H, Chen J, Shi M, et al. EDIL3 is a novel regulator of epithelial-mesenchymal transition controlling early recurrence of hepatocellular carcinoma. J Hepatol 2015;63:863-73. [Crossref] [PubMed]
- Zhang QY, Men CJ, Ding XW. Upregulation of microRNA-140-3p inhibits epithelial-mesenchymal transition, invasion, and metastasis of hepatocellular carcinoma through inactivation of the MAPK signaling pathway by targeting GRN. J Cell Biochem 2019;120:14885-98. [Crossref] [PubMed]
- Xiao S, Chang RM, Yang MY, et al. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology 2016;63:1256-71. [Crossref] [PubMed]
- Zeng S, Zhang Y, Ma J, et al. BMP4 promotes metastasis of hepatocellular carcinoma by an induction of epithelial-mesenchymal transition via upregulating ID2. Cancer Lett 2017;390:67-76. [Crossref] [PubMed]
- Yu J, Ren X, Chen Y, et al. Dysfunctional activation of neurotensin/IL-8 pathway in hepatocellular carcinoma is associated with increased inflammatory response in microenvironment, more epithelial mesenchymal transition in cancer and worse prognosis in patients. PLoS One 2013;8:e56069 [Crossref] [PubMed]
- Xiong C, Wang G, Bai D. A novel prognostic models for identifying the risk of hepatocellular carcinoma based on epithelial-mesenchymal transition-associated genes. Bioengineered 2020;11:1034-46. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)