
Bioinformatical identification of key genes regulated by IGF2BP2-mediated RNA N6-methyladenosine and prediction of prognosis in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is a malignant cancer with global prevalence, and its treatment remains unsatisfactory (1). While HCC has variable regional incidence, it nonetheless ranks as the fifth most common cause of cancer-related death worldwide. To date, most of the prognostic markers for HCC are derived from the postoperative period (2), and thus novel tissue or serum biomarkers to diagnose and predict HCC progress urgently need to be identified.
N6-methyladenosinde (m6A), as the most prevalent modification of messenger (mRNA) in eukaryotes, is part of a newly discovered mechanism of posttranscriptional gene regulation (3-5). It was reported that m6A occurs in the consensus sequences G (m6A) C or A (m6A) C (6), and more than 7,000 human genes could be m6A modified (4,5). As the dynamic modifier of mRNA, m6A is reversible. METTL3 and METTL14 work as the m6A methyltransferases (7-11); FTO or ALKBH5 play the role of demethylases (12,13); YTH domain family (including YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2) and insulin-like growth factor 2 mRNA binding proteins (IGF2BPs, including IGF2BP1/2/3) can recognize the m6A modified sites (14). M6A functions in the cellular process of RNA stability (11,15), translation efficiency (16,17), RNA secondary structure (18), subcellular localization (19), alternative polyadenylation, and splicing (20). In addition to cellular function, m6A methyltransferases are involved in the embryonic stem cell differentiation and circadian clock (19,21-23). Furthermore, m6A erasers are associated with human body weight (24-26) and mouse spermatogenesis (13).
m6A methylation has been found in cancer initiation and progression, and its dysregulation is associated with HCC. METTL3 is upregulated and contributes to HCC progression (27), while in metastatic HCC, decreased m6A modification has been observed (28). To date, the key target and biological function of m6A in HCC in HCC remain elusive.
Insulin-like growth factor 2 (IGF2) is a well-studied polypeptide growth factor, functioning in growth and development (29). Its mRNA binding proteins 1, 2, and 3 (IGF2BP1/2/3) function in mRNA stability and translation through m6A readers (30). In the past few years, abundant evidence has been produced showing the association of IGF2BP2 with type 2 diabetes and cancer (31). Autoimmune response to IGF2BP2 was observed in HCC, which is consistent with IGF2BP2 being functional in cancer progress regulation (32-34); however, the underlying mechanism linked to m6A modification remains to be explored.
In this study, we aim to identify tumor-associated genes regulated by IGF2BP2 on a m6A-dependent manner in HCC tissues using integrated bioinformatics analysis of RNA-sequencing of HCC patients in TCGA dataset (TCGA-LIHC) and m6A-sequencing/RNA-sequencing datasets of HCC cell line. We identified ten m6A-related IGF2BP2-regulated genes which are play key role in HCC progression. The relationship of the key genes, HCC patients’ clinical outcomes and immune infiltration were evaluated. Our results suggested that the key genes could be used as potential biomarkers for overall survival (OS) of HCC patients and act as an indicator for IGF2BP2 inhibition therapy in HCC.
I present the following article in accordance with the STREGA reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-306).
Methods
Data sets
The data of 369 HCC patients and 50 normal samples were obtained from TCGA-LICH database. The m6A-sequecning data (GSE90642), IGF2BP2-knockdown RNA-sequencing data (GSE80890 and GSE88139), and the gene expression microarray data of HCC or normal tissues (GSE102079) were downloaded from the Gene Expression Omnibus (GEO). The annotated human m6A sites data were obtained from RMBase (35). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
M6A-sequencing analysis
For M6A-sequencing data of HepG2 cells, the human genome GRCh38/hg38 was used to align the sequencing reads by HISAT2, and exomePeak was used to detect the m6A peaks.
Differentially expressed gene (DEG) identification
For IGF2BP2 knockdown RNA-sequencing, the human genome GRCh38/hg38 was used to align the sequencing reads by HISAT2 and assembled by StringTie, and DEGs were identified by DESeq2. For TCGA-LICH data, Gene Expression Profiling Interactive Analysis (GEPIA) was used for analysis. For microarray data of HCC, DEGs were identified by limma package in R language.
Gene ontology (GO) and pathway enrichment analysis
Metascape software was used for GO and pathway enrichment analysis.
Protein-protein interaction (PPI) network construction and module analysis
Search Tool for the Retrieval of Interacting Genes (STRING) combined with Cytoscape software were used for the construction of the PPI network. Gene list were dropped into STRING with a score >0.4 being considered as significant. The criteria were set as follows: MCODE scores >3 and number of nodes >4. Finally, function and pathway enrichment analysis were performed for genes in the modules.
Tumor microenvironment analysis
The composition of 22 immune cell subtypes in different groups of HCC tissues was determined by the CIBERSORT package (36). HCC tissues were classified according to the gene expression. The relationship between key modified, IGF2BP2-regulated and differentially expressed genes (MIRDEGs) and TIICs was determined by TIMER algorithm (37).
Statistical analysis
Differences were estimated using paired or unpaired Student’s t test depend on different analysis. A P-value <0.05 was considered statistically significant. Survival curves were obtained using the Kaplan-Meier method and compared using the log-rank test.
Results
Identification of m6A-modified and IGF2BP2-regulated DEGs in HCC
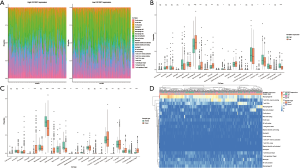
To determine the genes regulated by IGF2BP2 via a m6A-dependent manner in HCC, we first performed differential expression analysis between HCC tissues and normal tissues of the TCGA-LICH dataset. Then we explored the genes potentially regulated by IGF2BP2 by analyzing RNA-sequencing data of IGF2BP2-knockdown HepG2 cells. Furthermore, m6A-sequencing data of HepG2 cells was included in the analysis for filtering the genes with m6A modification. We screened 89 m6A-MIRDEGs in HCC tissues (Figure 1A).
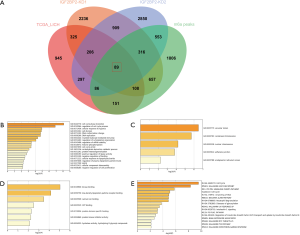
We speculated that most of these MIRDEGs were involved in HCC. To clarify the functions these genes were involved in, we uploaded all the MIRDEGs to Metascape to identify the overrepresented GO categories. Consistent with our previous assumptions, the most enriched MIRDEGs in biological process (BP) included cell cycle phase transition and regulation of cell cycle (Figure 1B). Moreover, the results showed that the MIRDEGs enriched in cell component (CC) were mainly involved in vacuolar lumen and chromosome (Figure 1C). The molecular function (MF) of MIRDEGs was enriched in kinase binding and low-density lipoprotein particle receptor binding (Figure 1D).
Next, we analyzed the most significant pathways of MIRDEGs functions integrated with Reactome, Canonical, BioCarta, Hallmark, and KEGG database using Metascape. The results indicated that the MIRDEGs were mostly enriched in cell cycle and Hallmark G2/m checkpoint (Figure 1E). Taken together, GO and pathway enrichment analysis indicated that the MIRDEGs might be involved in the progression of HCC by regulating HCC cell cycle.
Key candidate gene identification with PPI construction and modular analysis
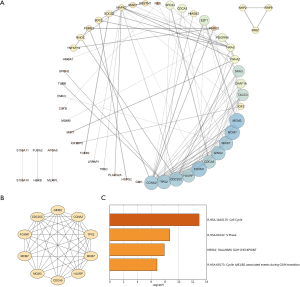
To further identify the genes playing key roles in HCC progression, a PPI network, containing 54 nodes and 121 edges, was constructed using the 89 MIRDEGs (Figure 2A). Among these, we chose a significant module from the PPI network complex using MCODE, which included MCM3, MCM7, MKI67, GINS2, CDCA5, FOXM1, HJURP, CDC25C, TPX2, and CCNA2 (Figure 2B). Surprisingly, we also found that this module was mainly associated with cell cycle (Figure 2C). These results further confirmed that MIRDEGs were involved in HCC progression through regulation of the cell cycle. Therefore, we named these 10 MIRDEGs as key MIRDEGs.
Validation of key MIRDEGs in independent datasets
To confirm the reliability and validity of the above analysis, we explored the expression profile of the key MIRDEGs in the expression microarray data of 152 HCC tissues and 14 normal liver tissues. Consistent with the previous analysis, the expression of these 10 genes were significantly different in tumors compared with normal tissues (Figure 3A,3B).
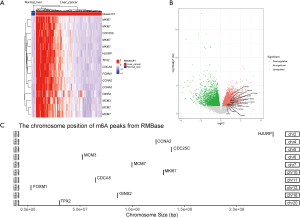
To confirm the presence of m6A modification in the mRNAs of the key MIRDEGs, we downloaded the processed m6A-sequencing and MeRIP-sequencing data from the RNA Modification Base (RMBase) database. As shown in Figure 3C, the m6A peaks were detected at the locus of the key MIRDEGs.
Taken together, we validated that the key MIRDEGs are containing m6A modification and play critical role in HCC progression.
Correlations between the key MIRDEGs and clinical features of HCC patients
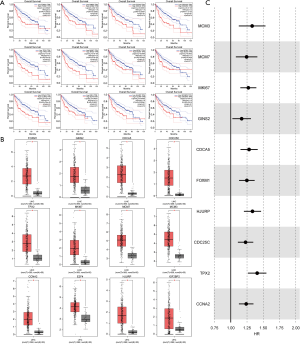
To further identify the clinical significance of the key MIRDEGs associated with HCC, we compared the overall survival (OS) between different groups of HCC patients from TCGA-LICH dataset classified by the key MIRDEGs expression level. Kaplan-Meier survival curves showed that patients with higher expression of the key MIRDEGs had shorter OS (Figure 4A). In addition, the key MIRDEGs were significantly upregulated in HCC tissues (Figure 4B). Cox regression analysis also showed that the expression of the key MIRDEGs related to OS (Figure 4C). Altogether, our results indicated that the key MIRDEGs are potential prognostic biomarkers for HCC patients.
The key MIRDEGs and HCC tumor microenvironment
As tumor-infiltrating immune cells (TIICs) have been reported to associated with the prognosis of multiple cancers, including HCC, we next estimate the infiltration of immune cells in TCGA-LIHC samples using CIBERSORT. We showed that the IGF2BP2 expression significantly changed variety of immune cells among TIICs subtypes (Figure 5A,5B). The infiltration of macrophages and lymphocytes is significantly different between IGF2BP2 high/low expression groups. However, we found that the immune cells change between HCC tissues and normal tissues is different (Figure 5C,5D). Using TIMER, we further showed that the tumor purity, B cells, CD8+ T cells, CD4+ T cells, Macrophages, Neutrophil and Dendritic cells were significantly associated with the expression of the key MIRDEGs (Figure 6).
Discussion
HCC is a highly aggressive global malignancy (38) and is responsible for approximately 700,000 deaths annually (39). It has a high mortality rate and low cure-to-incidence ratio (40). HCC’s late diagnosis is the primary cause of its low survival rate. In spite of the advances in surgery and patient care, only 10% of HCC patients are acceptable for resection and still suffer a significant risk of recurrence (39). The bleak treatment options highlight the necessity for the early detection of HCC (37). Due to HCC’s poor prognosis, there is an urgent need to develop new prognostic biomarkers. Using bioinformatical analysis, we identified 10 key m6A-modified genes that are regulated by IGF2BP2 in HCC, including MCM3, MCM7, MKI67, GINS2, CDCA5, FOXM1, HJURP, CDC25C, TPX2, and CCNA2, which named key MIRDEGs. Furthermore, we found that upregulation of the key MIRDEGs is associated with HCC prognosis. Hence, the key MIRDEGs might function as a novel set of prognostic biomarkers in HCC progression and indicator of IGF2BP2 inhibition therapy.
N6-methyladenosinde (m6A) is the most abundant internal mRNA modification and has an important role in RNA stability, translation efficiency, localization, splicing, and polyadenylation. m6A has diverse biological functions in cancer and is closely associated with several cancers types, and decreased m6A modification has been observed in HCC (28). Emerging evidence have suggested m6A abundance and the expression of its regulators including “writers”, “erasers” and “readers” are dysregulated in various types of cancers, including HCC, and they play critical roles in cancer initiation, progression, metastasis, as well as drug resistance and cancer relapse (41). Basically, the “writers” catalyze m6A modifications in the mRNA of oncogenes or tumor suppressor genes; and then the “readers” decoding the m6A methylations and generating the functional signal through a series of molecular biological effects, including regulation of translation, RNA splicing and stability, in turn upregulating oncogene expression or downregulating tumor suppressor gene expression. On the other hand, the “erases” can selectively remove m6A methylations from the mRNA of oncogenes or tumor suppressor genes, preventing the action of “readers”, thereby upregulating oncogene expression or downregulating tumor suppressor gene expression.
IGF2BP2 was originally identified as an IGF2 RNA-binding protein (RBP). It has since been discovered that it also targets the mRNA of LAMB2, LIMS2, TRIM54, UCP1, and another 12 other genes (42-45). One recent study suggested that, as a new m6A reader, RBP can recognize m6A modification and bind with m6A RNA through K homology (KH) domains (30), leading to the stability and upregulation of m6A-modified mRNA. Other research indicates that IGF2BP2 functions as a biomarkers in the screening, diagnosis, and prognosis of cancer, including HCC, colorectal, ovarian, and breast cancer (32,33,45). IGF2BP2 works as an oncogene in cancer (46-49). In 1999, the p62 isoform of IGF2BP2 was identified to be an autoantigen in HCC (50). In another study, 21% of HCC patients were found to have the autoantibody against p62, which was absent in chronic hepatitis and liver cirrhosis (50). Later, the overexpression of IGF2BP2 was identified in HCC (51), and later discoveries showed that high IGF2BP2 expression predicts poor prognosis in patients with HCC (52).
In this study, we conducted intergenic analysis of IGF2BP2 knockdown RNA-sequencing data, TCGA-LICH expression data, and m6A-sequencing data to identify IGF2BP2-regulated genes that function in m6A modification. A total of 89 IGF2BP2-related and m6A-modified genes were filtered. GO and pathway enrichment analysis indicated that these genes mainly enriched in cell cycle signaling pathway. Then we constructed a PPI network of the 89 gene candidates and extracted a significant module containing 10 key MIRDEGs from the PPI network, including MCM3, MCM7, MKI67, GINS2, CDCA5, FOXM1, HJURP, CDC25C, TPX2, and CCNA2. These key MIRDEGs were validated in an independent HCC microarray expression profile and an m6A database RMBase. To the best of our knowledge, these proto-oncogene genes have been explored in the prognosis of HCC. MCM3 and MCM7 play roles in DNA replication. In HCC, overexpressed translation regulator, Pokemon, was found to suppress the expression of MCM3 and MCM7, suggesting that MCM3 and MCM7 may be relevant to the prediction of HCC (53). Genome wide analysis in Southern China indicated that variants near TTN and CCDC8 were associated with markers of proliferation, MKI67, which could be potential biomarkers for the prediction of hepatitis B virus (HBV)-related HCC patients (54). Functioning in DNA replication, GINS complex subunit 2 (GINS2) was reported to be a novel prognostic cancer indicator, but its function in HCC has not been reported (55). Previously, CDCA5 was identified as a prognostic biomarker and therapeutic target for HCC (56), while downregulation of FOXM1 was shown to inhibit the proliferation, migration, and invasion of HCC cells (57). Furthermore, it has been discovered that the overexpression of Holliday junction-recognizing protein (HJURP) could promote proliferation of HCC cells (58). Meanwhile, dihydromyricetin (DHM) was found to induce G2/M phase arrest, which could lead to the inhibition of HCC progression through Chk1/Chk2/Cdc25C pathway. It was also revealed that TPX2 is involved in HCC cell proliferation, apoptosis, and epithelial-mesenchymal transition (EMT) (59). Finally, the expression of miR-22-CCNA2 has been associated with the proliferation of HCC (60). To our surprise, pathway enrichment analysis for the key MIRDEGs suggested that the key MIRDEGs might be involved in the HCC progression by regulating cell cycle pathway. The main characteristic of cancer is its relentless proliferation (61-64), which is usually regulated by a series of cell cycle regulators, including CycD1 (65,66), p21, p27, and p57 (67-69). This finding indicates that the key MIRDEGs have critical functions in HCC.
Although these genes have been investigated separately in HCC, their modification through m6A and IGF2BP2 regulation have not been examined as a whole. These 10 key genes could serve as a novel set of biomarkers to predict HCC progression. IGF2BP2 is the key to connecting all these genes with each other. As a novel m6A reader, IGF2BP2 may serve as the posttranscriptional bridge to link these key genes with HCC. Because the key MIRDEGs are regulated by IGF2BP2, it is considerable to use IGF2BP2 inhibition treatment when the dysregulation of key MIRDEGs was observed. In addition, we found that the key MIRDEGs expression was associated with the immune cell infiltration, suggesting that IGF2BP2 inhibition therapy may enhance the immune response to tumor cells, which demonstrating a potential HCC therapeutic strategy that IGF2BP2 inhibitor combined with checkpoint inhibitor (PD-1/PD-L1 inhibitors) to suppressing the HCC development and reducing the HCC recurrence after surgery.
In summary, we found that IGF2BP2 could recognize the 10 key MIRDEGs via m6A modification and stabilize their mRNA, which leads to the upregulation of these genes. This would disturb the normal cell cycle, alter the tumor microenvironment and promote HCC progression. Although the results remain need extensive experiment to validate, our study provides a new direction in HCC research and offers a novel set of biomarkers with potential prognostic and therapeutic significance.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The author has completed the STREGA reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-306
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-306). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Hwang HS, An J, Kang HJ, et al. Prognostic Molecular Indices of Resectable Hepatocellular Carcinoma: Implications of S100P for Early Recurrence. Ann Surg Oncol 2021; Epub ahead of print. [Crossref] [PubMed]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 1974;71:3971-5. [Crossref] [PubMed]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201-6. [Crossref] [PubMed]
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012;149:1635-46. [Crossref] [PubMed]
- Selmi T, Hussain S, Dietmann S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res 2021;49:1006-22. [Crossref] [PubMed]
- Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997;3:1233-47. [PubMed]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014;10:93-5. [Crossref] [PubMed]
- Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014;24:177-89. [Crossref] [PubMed]
- Tuck MT. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem J 1992;288:233-40. [Crossref] [PubMed]
- Wang X, He C. Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol 2014;11:669-72. [Crossref] [PubMed]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. [Crossref] [PubMed]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18-29. [Crossref] [PubMed]
- Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613 [Crossref] [PubMed]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. [Crossref] [PubMed]
- Meyer KD, Patil DP, Zhou J, et al. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell 2015;163:999-1010. [Crossref] [PubMed]
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. [Crossref] [PubMed]
- Liu N, Dai Q, Zheng G, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015;518:560-4. [Crossref] [PubMed]
- Fustin JM, Doi M, Yamaguchi Y, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013;155:793-806. [Crossref] [PubMed]
- Molinie B, Wang J, Lim KS, et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods 2016;13:692-8. [Crossref] [PubMed]
- Batista PJ, Molinie B, Wang J, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014;15:707-19. [Crossref] [PubMed]
- Wang Y, Li Y, Toth JI, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014;16:191-8. [Crossref] [PubMed]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015;347:1002-6. [Crossref] [PubMed]
- Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 2007;39:724-6. [Crossref] [PubMed]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007;316:889-94. [Crossref] [PubMed]
- Do R, Bailey SD, Desbiens K, et al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study. Diabetes 2008;57:1147-50. [Crossref] [PubMed]
- Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. [Crossref] [PubMed]
- Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017;65:529-43. [Crossref] [PubMed]
- Woźniak M, Duś-Szachniewicz K, Ziółkowski P. Insulin-Like Growth Factor-2 Is Induced Following 5-Aminolevulinic Acid-Mediated Photodynamic Therapy in SW620 Human Colon Cancer Cell Line. Int J Mol Sci 2015;16:23615-29. [Crossref] [PubMed]
- Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018;20:285-95. [Crossref] [PubMed]
- Bell JL, Wächter K, Mühleck B, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci 2013;70:2657-75. [Crossref] [PubMed]
- Liu W, Li Z, Xu W, et al. Humoral autoimmune response to IGF2 mRNA-binding protein (IMP2/p62) and its tissue-specific expression in colon cancer. Scand J Immunol 2013;77:255-60. [Crossref] [PubMed]
- Liu X, Ye H, Li L, et al. Humoral autoimmune responses to insulin-like growth factor II mRNA-binding proteins IMP1 and p62/IMP2 in ovarian cancer. J Immunol Res 2014;2014:326593 [Crossref] [PubMed]
- Liu W, Li Y, Wang B, et al. Autoimmune Response to IGF2 mRNA-Binding Protein 2 (IMP2/p62) in Breast Cancer. Scand J Immunol 2015;81:502-7. [Crossref] [PubMed]
- Sun WJ, Li JH, Liu S, et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res 2016;44:D259-65. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Lin Z, Cai YJ, Chen RC, et al. A microRNA expression profile for vascular invasion can predict overall survival in hepatocellular carcinoma. Clin Chim Acta 2017;469:171-9. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Huang H, Weng H, Deng X, et al. RNA Modifications in Cancer: Functions, Mechanisms, and Therapeutic Implications. Annual Review of Cancer Biology 2020;4:221-40. [Crossref]
- Nielsen J, Christiansen J, Lykke-Andersen J, et al. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 1999;19:1262-70. [Crossref] [PubMed]
- Boudoukha S, Cuvellier S, Polesskaya A. Role of the RNA-binding protein IMP-2 in muscle cell motility. Mol Cell Biol 2010;30:5710-25. [Crossref] [PubMed]
- Schaeffer V, Hansen KM, Morris DR, et al. RNA-binding protein IGF2BP2/IMP2 is required for laminin-β2 mRNA translation and is modulated by glucose concentration. Am J Physiol Renal Physiol 2012;303:F75-82. [Crossref] [PubMed]
- Dai N, Zhao L, Wrighting D, et al. IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and Other mRNAs encoding mitochondrial proteins. Cell Metab 2015;21:609-21. [Crossref] [PubMed]
- Barghash A, Helms V, Kessler SM. Overexpression of IGF2 mRNA-Binding Protein 2 (IMP2/p62) as a Feature of Basal-like Breast Cancer Correlates with Short Survival. Scand J Immunol 2015;82:142-3. [Crossref] [PubMed]
- Kessler SM, Laggai S, Barghash A, et al. IMP2/p62 induces genomic instability and an aggressive hepatocellular carcinoma phenotype. Cell Death Dis 2015;6:e1894 [Crossref] [PubMed]
- Barghash A, Golob-Schwarzl N, Helms V, et al. Elevated expression of the IGF2 mRNA binding protein 2 (IGF2BP2/IMP2) is linked to short survival and metastasis in esophageal adenocarcinoma. Oncotarget 2016;7:49743-50. [Crossref] [PubMed]
- Ye S, Song W, Xu X, et al. IGF2BP2 promotes colorectal cancer cell proliferation and survival through interfering with RAF-1 degradation by miR-195. FEBS Lett 2016;590:1641-50. [Crossref] [PubMed]
- Zhang JY, Chan EK, Peng XX, et al. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J Exp Med 1999;189:1101-10. [Crossref] [PubMed]
- Simon Y, Kessler SM, Bohle RM, et al. The insulin-like growth factor 2 (IGF2) mRNA-binding protein p62/IGF2BP2-2 as a promoter of NAFLD and HCC? Gut 2014;63:861-3. [Crossref] [PubMed]
- Kanda T, Goto T, Hirotsu Y, et al. Molecular Mechanisms Driving Progression of Liver Cirrhosis towards Hepatocellular Carcinoma in Chronic Hepatitis B and C Infections: A Review. Int J Mol Sci 2019;20:1358. [Crossref] [PubMed]
- Bi X, Jin Y, Gao X, et al. Investigation of Pokemon-regulated proteins in hepatocellular carcinoma using mass spectrometry-based multiplex quantitative proteomics. Eur J Mass Spectrom (Chichester) 2013;19:111-21. [Crossref] [PubMed]
- Yang CK, Yu TD, Han CY, et al. Genome-Wide Association Study of MKI67 Expression and its Clinical Implications in HBV-Related Hepatocellular Carcinoma in Southern China. Cell Physiol Biochem 2017;42:1342-57. [Crossref] [PubMed]
- Ouyang F, Liu J, Xia M, et al. GINS2 is a novel prognostic biomarker and promotes tumor progression in early-stage cervical cancer. Oncol Rep 2017;37:2652-62. [Crossref] [PubMed]
- Shen Z, Yu X, Zheng Y, et al. CDCA5 regulates proliferation in hepatocellular carcinoma and has potential as a negative prognostic marker. Onco Targets Ther 2018;11:891-901. [Crossref] [PubMed]
- Tian C, Wu H, Li C, et al. Downreguation of FoxM1 by miR-214 inhibits proliferation and migration in hepatocellular carcinoma. Gene Ther 2018;25:312-9. [Crossref] [PubMed]
- Hu B, Wang Q, Wang Y, et al. Holliday junction-recognizing protein promotes cell proliferation and correlates with unfavorable clinical outcome of hepatocellular carcinoma. Onco Targets Ther 2017;10:2601-7. [Crossref] [PubMed]
- Liang B, Jia C, Huang Y, et al. TPX2 Level Correlates with Hepatocellular Carcinoma Cell Proliferation, Apoptosis, and EMT. Dig Dis Sci 2015;60:2360-72. [Crossref] [PubMed]
- Yang F, Gong J, Wang G, et al. Waltonitone inhibits proliferation of hepatoma cells and tumorigenesis via FXR-miR-22-CCNA2 signaling pathway. Oncotarget 2016;7:75165-75. [Crossref] [PubMed]
- Brandi G, Calabrese C, Pantaleo MA, et al. Circadian variations of rectal cell proliferation in patients affected by advanced colorectal cancer. Cancer Lett 2004;208:193-6. [Crossref] [PubMed]
- Taniguchi H, Fernández AF, Setién F, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res 2009;69:8447-54. [Crossref] [PubMed]
- Yang X, Wood PA, Oh EY, et al. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat 2009;117:423-31. [Crossref] [PubMed]
- Tamai TK, Young LC, Cox CA, et al. Light acts on the zebrafish circadian clock to suppress rhythmic mitosis and cell proliferation. J Biol Rhythms 2012;27:226-36. [Crossref] [PubMed]
- Fu L, Pelicano H, Liu J, et al. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002;111:41-50. [Crossref] [PubMed]
- Dang CV. MYC on the path to cancer. Cell 2012;149:22-35. [Crossref] [PubMed]
- Massagué J. G1 cell-cycle control and cancer. Nature 2004;432:298-306. [Crossref] [PubMed]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9:400-14. [Crossref] [PubMed]
- Herkert B, Eilers M. Transcriptional repression: the dark side of myc. Genes Cancer 2010;1:580-6. [Crossref] [PubMed]


