
Screening of a novel autophagy-related prognostic signature and therapeutic targets in hepatocellular carcinoma
Introduction
Liver cancer is the fifth leading cause of all malignant tumors and the third leading cause of cancer-related mortality (1,2). According to the latest statistics, it has been estimated that new cases may reach 42,230 in 2021, with nearly 30,230 deaths in the United States (3). The incidence and mortality of liver cancer are increasing yearly. Hepatocellular carcinoma (HCC) accounts for 85% of all liver cancers, which is a highly heterogeneous tumor (4,5). As previously reported, HCC is highly associated with chronic hepatitis B virus (HBV) or HCV infection, exposure to aflatoxins, nonalcoholic fatty liver disease (NAFLD), as well as alcohol abuse (6,7). Although intensive efforts have been made to treat HCC, including surgical resection, liver transplantation, vascular intervention, or radiofrequency ablation, the overall survival (OS) of HCC has remained unsatisfactory and the 5-year survival rate is approximately 18% in the United States (8-10). As a result, there is an urgent need to construct a useful prognostic model to classify patients at high risk of recurrence or distal metastasis. As previously reported, conventional models have been established by integrating tumor-node-metastasis (TNM) stages, tumor grades, age, clinical pathological stages, or vascular invasion to predict the prognosis of HCC (11-13). Nevertheless, the overall predictive efficiency of constructed models is still limited, owing to the remarkable heterogeneity of HCC (14,15). Therefore, it is of great significance to elucidate the underlying molecular mechanisms of HCC progression and identify novel biomarkers to improve the outcomes and prognosis prediction of HCC patients.
Autophagy is a well-known metabolic crosstalk that maintains the intra-cellular homeostasis via a lysosomal degradation pathway (16-18). Previous knowledge has indicated that autophagy is indispensable for cell survival, differentiation, and development (19). Autophagy could suppress tumorigenesis in the early stages via eliminating damaged proteins and organelles to attenuate chromosomal instability (20,21). However, autophagy can also enhance tumor progression through supplying nutritional contents for tumor survival under some stresses, like hypoxia, ischemia, or cell starvation (18,22). We thus intended to identify abnormal autophagy-associated genes to construct nomogram in HCC. Mutations of autophagy-related genes (ARGs), like ATG2B, ATG4, ATG5, and ATG12, were found to be associated with multiple cancers, like familial myeloid malignancies, gastric, and colorectal cancers (23,24). Previous study reported that the expression of long non-coding (lnc)-HOTAIR and ATG-7 are significant to predict the development and prognostic outcomes of HCC patients with HCV-4 infection (25). Besides, Yu et al. also indicated that miR-155-5p could reverse adriamycin resistance in HCC by targeting ATG5 (26). These ATGs might be modulated by several levels of crosstalk and in turn regulate multiple signaling pathways, which could be targetable with useful drugs. As a result, how to identify ARGs associated with HCC prognosis and progression is meaningful for further management of HCC.
In the current study, owing to the availability and development of high-throughput sequencing data, it was feasible for us to systematically assess the associations between ARGs and the survival prognosis of HCC patients based on these public datasets. We intended to identify and verify an ARGs signature that could predict prognosis and provide therapeutic vulnerabilities in HCC. Different from previous studies, we also illustrated the overall gene expression profiles and discussed the relationships between ARGs and immune infiltrations. We further provided a novel nomogram model for HCC patients with remarkable clinical significance. We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-664).
Methods
Identification of differentially expressed genes (DEGs)
The raw gene expression levels of resistance to sorafenib in HCC were downloaded from GSE94550 database. The limma package in R (https://bioconductor.org/packages/release/bioc/html/limma.html) was used to obtain the DEGs with more than a 1.5-fold difference in expression (adj.P<0.01). Moreover, we obtained the genes involving in autophagy processes from the Molecular Signatures Database (MSigDB) and National Center for Biotechnology Information (NCBI)-Gene website by using the keyword “autophagy”. The candidate genes were used to further analyze intersection with DEGs and ARGs. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Functional enrichment analysis
Functional enrichment analysis of autophagy-related DEGs was performed by clusterProfiler R package (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) to identify Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with P<0.05 were used as the cut-off. The visualization was performed using ggplot2 package in R software (https://cran.r-project.org/web/packages/ggplot2/index.html).
Establishment of experimental model
Univariate Cox analysis was used to initially identify potential prognostic candidate genes (27). Least absolute shrinkage and selection operator (LASSO) penalty Cox regression analysis was used for confirmation (28). Based on the optimal lambda value that was selected through 1,000 cross-validations, a panel of prognostic candidate genes was determined. The regression coefficient of the Cox regression model was used to calculate the risk scores by using The Cancer Genome Atlas (TCGA)-Liver Hepatocellular Carcinoma (LIHC) database. Risk score = (Coefficientmgene_1 × gene_1 expression) + (Coefficientmgene_2 × gene_2 expression) + … + (Coefficientgene_n × gene_n expression) (29). With the median risk score as cutoff, all participants in the training data were split into a high-risk group and a low-risk group. International Cancer Genome Consortium (ICGC) data were used as testing data for validation. The survival curve with high-risk and low-risk was analyzed by Kaplan-Meier survival analysis and the log-rank test. Nomogram-based clinical modeling has been one of the most widely used statistic methods in clinical investigations. A receiver operating characteristic (ROC) curve was used for the sensitivity and specificity of the nomogram.
Immune score determination for the microenvironment
The gene expression amounts of prognostic autophagy-related DEGs were normalized using the limma package. Then, immune score stromal scores were calculated by applying the Estimation of Stromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm to the normalized matrix data (30). The scores were used to predict the infiltration of non-tumor cells, by analyzing the specific gene expression signature of immune and stromal cells (30).
Single nucleotide polymorphism (SNP) analysis
TCGA SNP data were downloaded from the Genome Data Commons Data Portal (https://portal.gdc.cancer.gov). The maftools R package (https://bioconductor.org/packages/release/bioc/html/maftools.html) provided a multitude of analysis modules to perform the visualization process (31).
Statistical analysis
The survival package was used to perform the Cox regression model. The normalization and differentially expressed analysis were conducted using the limma package. All statistical analysis was implemented based on the R software (R 4.1.0; https://cran.r-project.org/bin/windows/base/). A P value <0.05 was considered as significant.
Results
Identification of ARGs
Firstly, we searched the ARGs via key word of autophagy from NCBI-Gene and MSigDB datasets. The list of ARGs was downloaded with the keyword of autophagy from the NCBI-Gene. Besides, we also accessed the MSigDB dataset via the identity email to obtain the ARGs. The final ARGs derived from the two datasets were overlapped for the following study, and are summarized in https://cdn.amegroups.cn/static/public/jgo-21-664-1.xlsx.
Screening of differentially expressed ARGs (DE-ARGs)
To identify the DE-ARGs in HCC, we conducted the differential analysis based on the limma package. We obtained the transcriptome matrix data from the GSE94550 dataset, including 6 HCC samples and 3 normal control samples. The expression data of ARGs was extracted and differential analysis was performed then with the threshold of adj.P.Val <0.05 and |logFC| >1.0. As shown in Figure 1A, 856 sorafenib resistance-related DEGs were identified in the volcano plot. Subsequently, integration analysis was conducted with the DE-ARGs to finally find the 106 differential SRGs, of which 61 genes were up-regulated and 45 were down-regulated. The results were further illustrated in Venn diagram and heatmap via ggplot2 package (Figure 1B,1C). In order to further analyze the relationships between ARGs and HCC survival, 28 genes with significant differences were identified based on the univariate Cox regression analysis with P<0.05 (Figure 1D). In addition, Kaplan-Meier analysis was performed to identify 13 ARGs that correlated with the OS of HCC. We thus integrated the results from the univariate Cox regression analysis and Kaplan-Meier analysis to overlap the final 10 candidates (Figure 1E).
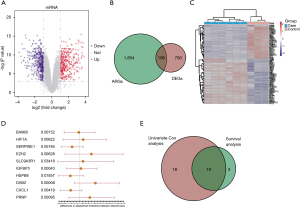
GO and KEGG analysis based on DE-ARGs
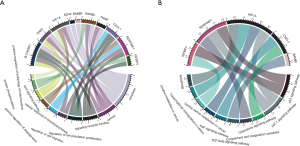
To analyze and ascertain the functional and biological significance of altered ARGs in HCC, we utilized the clusterProfiler package to conduct GO and KEGG analysis, which were visualized in chord diagrams via ggplot2. The results indicated that dysregulated DE-ARGs were mainly involved in regulation of autophagy of mitochondrion, Wnt signaling pathway, cell population proliferation, and cell migration biological items (Figure 2A). The significantly enriched pathways mainly contained autophagy, transcriptional misregulation in cancer, transforming growth factor-β (TGF-β), tumor necrosis factor (TNF), chemokine signaling pathway, and complement and coagulation cascades crosstalk (Figure 2B).
Assessment of predictive efficiency of ARGs model
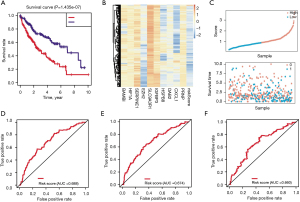
Based on the results from the Cox regression and Kaplan-Meier analyses, we constructed the predictive model using the 10 candidates and calculated the corresponding risk scores. The risk formula was calculated as the following: risk scores = expression of gene1 × (coefficient of gene1) + the expression of gene2 × (coefficient of gene2) + … + expression of gene10 × (coefficient of gene10), where Coefficient meant the coefficient of the gene within the prognostic model. Kaplan Meier analysis then indicated that HCC patients with high-risk scores had a lower survival rate (log-rank test P=1.435e-07; Figure 3A). As shown in Figure 3B, the heatmap revealed the expression data of genes in the risk model. As indicated in Figure 3C, the scatter plot of the risk scores and survival time across the samples were illustrated. The ROC curve analysis revealed that the ARGs risk model had a well diagnostic positive rate [1-year area under the curve (AUC) =0.688; Figure 3D], and the 3- and 5-year AUCs were 0.674 and 0.66, respectively (Figure 3E,3F).
Validation of ARGs predictive model in the external HCC datasets
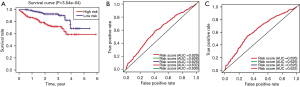
To further verify the predictive accuracy of established model, we utilized the ICGC-HCC cohort as the external dataset to perform the validations. The median risk score was used as the cutoff to classify the samples into high- and low-risk groups. Additionally, Kaplan-Meier analysis also suggested that samples with high-risk scores had lower survival rates relative to those with low-risk scores (log-rank test P<0.0001; Figure 4A). Lastly, the 1- and 3-year AUC for predicting false positive diagnostic effects were 0.624 and 0.635, respectively (Figure 4B,4C).
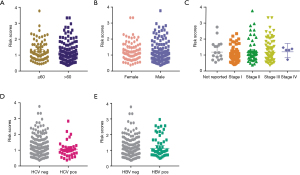
Differential analysis of ARGs scores across the clinical characteristics of HCC
To further discuss the differences of ARGs in clinical factors, we collected the clinical information of HCC samples and merged them with ARGs scores into one matrix. Subsequently, correlation analysis indicated that only advanced tumor stages was positively associated with high ARGs scores with P=0.0227 (Figure 5A-5E). However, no significant associations were found between ARGs with other clinical parameters, including age (P=0.9306), gender (P=0.4325), HBV infection (P=0.3573), and HCV infection (P=0.1243).
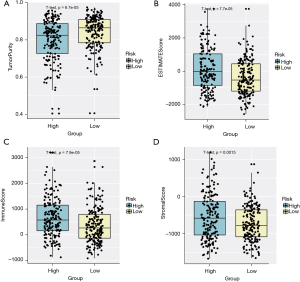
Assessment of ImmuneScore, StromalScore, and tumor purity in high- and low-risk HCC samples
We utilized the ESTIMATE algorithm to analyze the differences of ImmuneScore, StromalScore, and tumor purity in high- and low-risk samples. In the HCC tumor microenvironment (TME), immune cells and stromal cells are the two main types of cells. The ESTIMATE algorithm was carried out based on the transcriptome data to calculate the ImmuneScore and StromalScore for each sample, thereby inferring the proportions of the two types of cells. Samples with high infiltrating levels of immune cells and stromal cells often have low levels of tumor purity (32,33). As shown in Figure 6, there were significant differences of tumor purity (P=6.71e-05), infiltrating cell analysis (P=7.77e-05), immune analysis (P=7.9e-05), and stromal cells analysis (P=0.0015). We thus speculated that high-risk samples may have disorders of immune regulations.
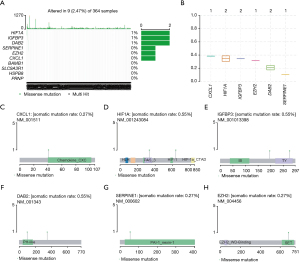
Differential mutation profiles of ARGs in high- and low-risk HCC samples
To analyze the mutation information of ARGs in high- and low-risk groups, we downloaded the maf profiles of TCGA-LIHC and used the maftool package to evaluate the SNP information. Among the genes within the ARGs risk model, we found that 6 genes had notable mutation information, in which HIF1A, IGFBP3, and DAB2 had high frequent missense mutations (Figure 7A). Besides, the results of variant allele frequency indicated that CXCL1 had the highest variant frequency (Figure 7B). Lastly, we utilized the lollipopPlot (https://www.rdocumentation.org/packages/maftools/versions/0.99.30/topics/lollipopPlot) to visualize the SNP information of each gene, including CXCL1, HIF1A, IGFBP3, DAB2, SERPINE1, and EZH2 (Figure 7C-7H).
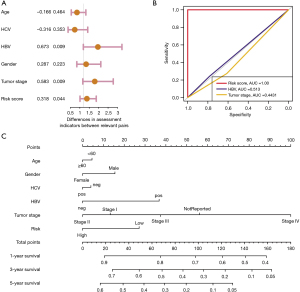
Independent prognostic analysis of ARGs in HCC
To analyze the relationships between ARGs scores with clinical characteristics in HCC, univariate Cox regression analysis was conducted to evaluate the regression associations between clinical factors and OS of HCC. Univariate Cox regression results indicated that HBV infection (HR =0.673; P=0.009), tumor stage (HR =0.583; P=0.009), and clinical factors and ARGs risk scores (HR =0.318; P=0.044) were all significantly related to OS of HCC (Figure 8A). The ROC analysis indicated that ARGs risk scores had the highest predictive efficiency with AUC =1.0, followed by HBV infection with AUC =0.513, and tumor stage with AUC =0.443 (Figure 8B). Lastly, we also established the nomogram to predict OS of HCC by integrating ARGs scores and clinical parameters, where the points in the first arrow represented the score references for each variable. We thus calculated the predictive scores of each variable for each participant and summed them to obtain the overall scores. The sum of scores were mapped into the total points to obtain the 1-, 3-, and 5-year of predictive OS probabilities (Figure 8C).
Discussion
As a highly aggressive cancer, HCC is prone to recurrence and to having worse outcomes (34-36). Therefore, diagnostic and therapeutic biomarkers with high predictive accuracy are urgently warranted. Some researchers have observed that autophagy is highly related to distal metastasis, drug resistance, and progression of HCC (4,37,38). However, intensive studies have mainly given attention to the impacts of ARGs in tumorigenesis and drug development, whereas few studies have addressed the prognostic significance of ARGs in cancer. In recent years, many studies have utilized the m6A-related genes, glycolysis-related genes, hypoxia-related genes, or ferroptosis-related genes to establish prognostic signatures, respectively, to predict the prognosis of HCC (39,40). Lin et al. previously constructed a novel nomogram that predicts OS in HCC patients (41). However, this study did not further discuss the differential mutation profiles of ARGs and immune infiltration patterns in high- and low-risk ARGs groups. Besides, owing to different screening procedures, the DE-ARGs and results in the two studies are also distinct.
Here, we downloaded the expression data of ARGs of HCC patients from TCGA dataset. First of all, we conducted differential analysis between the tumor samples and normal tissues to identify a total of 106 DEGs. Functional enrichment analysis suggested that these DEGs were mainly enriched in autophagy of mitochondrion, Wnt signaling pathway, cell population proliferation, and cell migration biological items. Afterwards, the ARGs risk scores were calculated based on Cox regression analysis by integrating the 10 genes into the following formula: risk scores = expression of gene1 × (coefficient of gene1) + the expression of gene2 × (coefficient of gene2) + … + expression of gene10 × (coefficient of gene10). To further validate the predictive accuracy of established model, we utilized the ICGC-HCC cohort as the external dataset to perform the validations. We also utilized the ESTIMATE algorithm to analyze the differences of ImmuneScore, StromalScore, and tumor purity in high- and low-risk samples. Among the genes within the ARGs risk model, we found that 6 genes had notable mutation information, among which HIF1A, IGFBP3, and DAB2 had high frequent missense mutations. Lastly, we also established a nomogram to predict OS of HCC by integrating ARGs scores and clinical parameters, which has great clinical significance. Compared with other nomograms that were established based on other molecular signature, we further discussed the underlying relationships between ARGs and immune infiltrating cells and mutation profiles. We thus speculated that our established nomogram might have the capacity to predict the immunological responses and drug efficiency in HCC patients, which was interesting and meaningful in the following studies.
In this study, we used the ESTIMATE algorithm to find the differences of ImmuneScore, StromalScore, and tumor purity in high- and low-risk samples (42-44). We found that samples with high-risk ARGs have high infiltrations of immune cells and low levels of tumor purity. As previously reported, autophagy has been reported to participate in immune regulations, including tumor-associated macrophages, dendritic cells (DCs), and T or B lymphocytes (45,46). Besides, the release of cytokines from tumor or immune cells were also tightly influenced by autophagy (47). Mutually, some cytokines or immune cells also have effects on the function of autophagy, including interleukin (IL)-12, TGF-β, and interferon (IFN)-γ (48,49). Considering that immunotherapy has become an effective strategy to inhibit HCC, there is an urgent need to deeply determine the underlying associations between autophagy activation and efficacy of immune checkpoint inhibitors. Cen et al. revealed that autophagy deficiency could promote triple-negative breast cancer resistance to T cell-mediated cytotoxicity via blocking tenascin-C degradation (50). Also, autophagy inhibition upregulates CD4+ tumor infiltrating lymphocyte expression via miR-155 regulation and TRAIL activation (51). These results indicate that autophagy inhibition could prevent tumor associated immunosuppression and promote immune cell infiltration in TME, therefore providing a potential therapeutic strategy to inhibit progression and metastasis of tumors. We also found that HIF1A, IGFBP3, and DAB2 have high frequent missense mutations in high-risk ARGs groups. Previous study indicated that HIF1A and NFAT5 could coordinate Na+-boosted antibacterial defense via enhanced autophagy and autolysosomal targeting (52). Hypoxia-induced autophagy could also enhance colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway (53). However, no relative studies have clearly elucidated the relationship between HIF1A mutations and HCC progression.
However, this study involved several limitations that require further improvement. First of all, due to the limited cases in the study, we should collect more samples to validate the efficacy and robustness of the established ARGs signature model. Besides, the overall predictive efficiency of the nomogram is still limited. How to integrate clinical parameters and ARGs scores in HCC is meaningful to ultimately optimizing the model. Lastly, experimental assays are also needed to clarify the underlying associations between selected ARGs and HCC progression.
Taken together, our study established a robust autophagy-related risk score model to predict the prognosis of HCC, providing therapeutic targets. Meanwhile, we also discussed the differential immune infiltrations and mutation profiles between high- and low-risk ARGs samples. Last of all, we constructed a novel prognostic nomogram incorporating both the ARGs and clinical factors for providing individualized survival prediction.
Acknowledgments
Funding: The study was supported by The Guangzhou Science and Technology Program Guangzhou Science and Technology Planning Project (202102080618); The Guangdong Basic and Applied Basic Research Foundation (2015A030313101); Grant from Guangdong Science and Technology Department (2015B050501004, 2017B030314026); The Special Research Foundation of National Nature Science Foundation of China (81900285,81972262); Grant [2013]163 from Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology and Grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-664
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-664). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Losic B, Craig AJ, Villacorta-Martin C, et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat Commun 2020;11:291. [Crossref] [PubMed]
- Yu J, Green MD, Li S, et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med 2021;27:152-64. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Craig AJ, von Felden J, Garcia-Lezana T, et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2020;17:139-52. [Crossref] [PubMed]
- Zheng H, Pomyen Y, Hernandez MO, et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 2018;68:127-40. [Crossref] [PubMed]
- Villanueva A. Hepatocellular carcinoma. N Engl J Med 2019;380:1450-62. [Crossref] [PubMed]
- Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology 2019;156:477-91.e1. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, et al. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol 2016;25:74-85. [Crossref] [PubMed]
- Yoh T, Seo S, Taura K, et al. Surgery for recurrent hepatocellular carcinoma: achieving long-term survival. Ann Surg 2021;273:792-9. [Crossref] [PubMed]
- Liang JY, Wang DS, Lin HC, et al. A novel ferroptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Int J Biol Sci 2020;16:2430-41. [Crossref] [PubMed]
- Zhang B, Tang B, Gao J, et al. A hypoxia-related signature for clinically predicting diagnosis, prognosis and immune microenvironment of hepatocellular carcinoma patients. J Transl Med 2020;18:342. [Crossref] [PubMed]
- Li W, Chen QF, Huang T, et al. Identification and validation of a prognostic lncRNA signature for hepatocellular carcinoma. Front Oncol 2020;10:780. [Crossref] [PubMed]
- Xu LX, He MH, Dai ZH, et al. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol 2019;30:990-7. [Crossref] [PubMed]
- Yin Z, Dong C, Jiang K, et al. Heterogeneity of cancer-associated fibroblasts and roles in the progression, prognosis, and therapy of hepatocellular carcinoma. J Hematol Oncol 2019;12:101. [Crossref] [PubMed]
- Piffoux M, Eriau E, Cassier PA. Autophagy as a therapeutic target in pancreatic cancer. Br J Cancer 2021;124:333-44. [Crossref] [PubMed]
- Li W, He P, Huang Y, et al. Selective autophagy of intracellular organelles: recent research advances. Theranostics 2021;11:222-56. [Crossref] [PubMed]
- Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer 2020;19:12. [Crossref] [PubMed]
- Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol 2019;19:170-83. [Crossref] [PubMed]
- Mowers EE, Sharifi MN, Macleod KF. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J 2018;285:1751-66. [Crossref] [PubMed]
- Nassour J, Radford R, Correia A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 2019;565:659-63. [Crossref] [PubMed]
- Onorati AV, Dyczynski M, Ojha R, et al. Targeting autophagy in cancer. Cancer 2018;124:3307-18. [Crossref] [PubMed]
- Kang MR, Kim MS, Oh JE, et al. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol 2009;217:702-6. [Crossref] [PubMed]
- Saliba J, Saint-Martin C, Di Stefano A, et al. Germline duplication of ATG2B and GSKIP predisposes to familial myeloid malignancies. Nat Genet 2015;47:1131-40. [Crossref] [PubMed]
- Feng X, Zhang H, Meng L, et al. Hypoxia-induced acetylation of PAK1 enhances autophagy and promotes brain tumorigenesis via phosphorylating ATG5. Autophagy 2021;17:723-42. [Crossref] [PubMed]
- Yu Q, Xu XP, Yin XM, et al. miR-155-5p increases the sensitivity of liver cancer cells to adriamycin by regulating ATG5-mediated autophagy. Neoplasma 2021;68:87-95. [Crossref] [PubMed]
- Wen F, Huang J, Lu X, et al. Identification and prognostic value of metabolism-related genes in gastric cancer. Aging (Albany NY) 2020; Epub ahead of print. [Crossref] [PubMed]
- Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997;16:385-95. [Crossref] [PubMed]
- Zhang Y, Li H, Zhang W, et al. LASSO-based Cox-PH model identifies an 11-lncRNA signature for prognosis prediction in gastric cancer. Mol Med Rep 2018;18:5579-93. [Crossref] [PubMed]
- Zhang C, Zheng JH, Lin ZH, et al. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of osteosarcoma. Aging (Albany NY) 2020;12:3486-501. [Crossref] [PubMed]
- Shah N, Wang P, Wongvipat J, et al. Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. Elife 2017;6:27861. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Dai T, Peng L, Lin G, et al. Preoperative elevated plasma fibrinogen level predicts tumor recurrence and poor prognosis in patients with hepatocellular carcinoma. J Gastrointest Oncol 2019;10:1049-63. [Crossref] [PubMed]
- Qin W, Han C, Mai R, et al. Establishment of a prognostic model for predicting short-term disease-free survival in cases of hepatitis B-related hepatocellular carcinoma with the TP53 249Ser mutation in southern China. Transl Cancer Res 2020;9:4517-33. [Crossref]
- Ahn JC, Teng PC, Chen PJ, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology 2021;73:422-36. [Crossref] [PubMed]
- Yu S, Wang Y, Jing L, et al. Autophagy in the "inflammation-carcinogenesis" pathway of liver and HCC immunotherapy. Cancer Lett 2017;411:82-9. [Crossref] [PubMed]
- Lin Z, Niu Y, Wan A, et al. RNA m6 A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J 2020;39:e103181. [Crossref] [PubMed]
- Zhang Z, Guo M, Li Y, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy 2020;16:1482-505. [Crossref] [PubMed]
- Lu C, Fang S, Weng Q, et al. Integrated analysis reveals critical glycolytic regulators in hepatocellular carcinoma. Cell Commun Signal 2020;18:97. [Crossref] [PubMed]
- Lin Y, Wei X, Jian Z, et al. METTL3 expression is associated with glycolysis metabolism and sensitivity to glycolytic stress in hepatocellular carcinoma. Cancer Med 2020;9:2859-67. [Crossref] [PubMed]
- Fang Q, Chen H. Development of a novel autophagy-related prognostic signature and nomogram for hepatocellular carcinoma. Front Oncol 2020;10:591356. [Crossref] [PubMed]
- Ni J, Liu S, Qi F, et al. Screening TCGA database for prognostic genes in lower grade glioma microenvironment. Ann Transl Med 2020;8:209. [Crossref] [PubMed]
- Xu M, Li Y, Li W, et al. Immune and stroma related genes in breast cancer: a comprehensive analysis of tumor microenvironment based on the Cancer Genome Atlas (TCGA) database. Front Med (Lausanne) 2020;7:64. [Crossref] [PubMed]
- Liu Y, Wu J, Huang W, et al. Development and validation of a hypoxia-immune-based microenvironment gene signature for risk stratification in gastric cancer. J Transl Med 2020;18:201. [Crossref] [PubMed]
- Shibutani ST, Saitoh T, Nowag H, et al. Autophagy and autophagy-related proteins in the immune system. Nat Immunol 2015;16:1014-24. [Crossref] [PubMed]
- Germic N, Frangez Z, Yousefi S, et al. Regulation of the innate immune system by autophagy: neutrophils, eosinophils, mast cells, NK cells. Cell Death Differ 2019;26:703-14. [Crossref] [PubMed]
- Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy 2018;14:243-51. [Crossref] [PubMed]
- Yang R, Yang E, Shen L, et al. IL-12+IL-18 cosignaling in human macrophages and lung epithelial cells activates cathelicidin and autophagy, inhibiting intracellular mycobacterial growth. J Immunol 2018;200:2405-17. [Crossref] [PubMed]
- Cen S, Wang P, Xie Z, et al. Autophagy enhances mesenchymal stem cell-mediated CD4+ T cell migration and differentiation through CXCL8 and TGF-β1. Stem Cell Res Ther 2019;10:265. [Crossref] [PubMed]
- Li ZL, Zhang HL, Huang Y, et al. Autophagy deficiency promotes triple-negative breast cancer resistance to T cell-mediated cytotoxicity by blocking tenascin-C degradation. Nat Commun 2020;11:3806. [Crossref] [PubMed]
- Zarogoulidis P, Petanidis S, Domvri K, et al. Autophagy inhibition upregulates CD4+ tumor infiltrating lymphocyte expression via miR-155 regulation and TRAIL activation. Mol Oncol 2016;10:1516-31. [Crossref] [PubMed]
- Neubert P, Weichselbaum A, Reitinger C, et al. HIF1A and NFAT5 coordinate Na+-boosted antibacterial defense via enhanced autophagy and autolysosomal targeting. Autophagy 2019;15:1899-916. [Crossref] [PubMed]
(English Language Editor: J. Jones)


