
Utility of tumor-informed circulating tumor DNA in the clinical management of gastrointestinal malignancies
Introduction
Gastrointestinal (GI) and hepatobiliary/pancreatic malignancies account for approximately 26.7% of all global cancers, but more importantly account for 36.5% of the global cancer related mortality (1). Long-term survival is achieved mainly in patients with early-stage disease. A substantial number of patients affected by these neoplasms may not have early presentation or receive timely treatment. In addition to surgical resection, (neo)adjuvant systemic treatment is recommended by guidelines to minimize the risk of recurrence (2-4). The main objective of adjuvant chemotherapy in GI malignancies is to eradicate microscopic spread of the tumor at the time of diagnosis but are present after curative intent surgical resection, which by definition is not detected on routine imaging (e.g., computed tomography) or based on traditional tumor markers (CEA, CA 19-9, AFP, etc.) (5,6). This state is also known as molecular residual disease (MRD) (6). However, all currently available histopathologic prognostic factors to guide adjuvant treatment are based on the probability of MRD, rather than actual detection of MRD. In other words, pathologic staging, in addition to ‘high risk’ features such as angiolymphatic and perineural invasion predict the probability of MRD for patients with a defined stage in general, but for the individual patient they can lead to over- or undertreatment since they cannot identify the actual MRD state for an individual patient (7).
Liquid biopsy assays detect circulating tumor DNA (ctDNA), short fragments of DNA released by tumor cells. Conventional static assays seek genomic alterations in a set number of pre-defined cancer-associated genes, independent of tumor tissue (6). While they have a high rate of specificity, and can guide treatment decisions in advanced disease based on detection of validated predictive markers, their sensitivity is dependent on tumor stage, i.e., disease volume. Thus, static assays have limited utility for the detection of MRD after surgical removal of all visible disease. On the other hand, a tumor-informed ctDNA assay first identifies 16 tumor unique truncal mutations, and then uses a custom multiplex PCR assay to detect these 16 mutations in the plasma of patients for the presence of residual tumor (8,9). Using this approach, the sensitivity for MRD detection is improved by at least an order of magnitude compared to static assays. Furthermore, in addition to the tumor tissue, the patient’s blood is collected to compare with the tumor-derived mutations, excluding any potential clonal hematopoiesis of indeterminate potential (CHIP) mutations, which further increases the specificity of the assay (10). While the prognostic value of a positive MRD assay has been shown in initial studies for multiple tumor types (e.g., lung, bladder, breast), the largest body of evidence for GI cancers currently is for colorectal carcinomas (11-13).
Here, we sought to expand on currently available data for the role of tumor-informed ctDNA testing in a variety of GI malignancies, specifically with a focus on real world application of the test and inclusion of ethnic minorities which might be underrepresented in other published series. We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-484).
Methods
This was a retrospective chart review. Included patients were 18 years and older with a diagnosis of one of the following GI malignancies: cancers of the esophagus, stomach, small bowel, colon or rectum, anus, primary liver, bile ducts or pancreas. Patients with unknown primary cancer were excluded from this study. A personalized and tumor-informed multiplex PCR assay (SignateraTM bespoke mPCR NGS assay) was used for the detection and quantification of ctDNA. The technical details have been described previously (8). In brief, tumors underwent whole-exome sequencing, and 16 high-ranked patient-specific somatic single-nucleotide variants and short indels were selected for each patient. Multiplex PCR primer pairs were generated based on the selected set of variants. Cell-free DNA was extracted from plasma, universal libraries were created by end repair, A-tailing, and ligation with custom adapters. The libraries were then amplified by multiplex PCR, barcoded, pooled, and sequenced on an NGS sequencing platform (HiSeq 2500 system, Illumina Inc.). Plasma samples with two or more variants detected were defined as ctDNA positive. The first plasma sample for all patients with Stage I-III cancers was drawn >4 weeks after curative intent surgery. For Stage IV cancers, surgery had either been performed previously for early stage disease, or for curative intent in those patients with oligometastatic disease. Serial time points were collected on a subset of patients to monitor their ctDNA levels in response to treatment. All patients included had at least one available bespoke ctDNA test result between October 2019 and February 2021.
Statistical analysis
Following data were abstracted via chart review from eligible patients: age, gender, ethnicity, cancer stage, diagnosis date, surgical date, ctDNA status, microsatellite instability, tumor grade, and treatment. Data were summarized in tables. Chi square test was used to compare rate of ctDNA positivity between groups; P<0.05 was considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The current study was approved by the Institutional Review Board at the University of California Irvine (protocol# 2020-6196) and Saint Luke’s Health System (protocol# 001CTDNA21), and individual consent for this retrospective anonymized study was waived.
Results
Patient and tumor characteristics
Our study consisted of a total of 258 plasma samples from 198 patients. Table 1 depicts the patient and tumor characteristics. Sixty one percent were female, and despite a median age of 62 years, a wide range of ages were included (23–89 years). Slightly more than half of the patients were Caucasian, while 49% belonged to ethnic minorities (Asian, Hispanic, and African American) and 4% identified as ‘other’.
Table 1
| N | (%) | |
|---|---|---|
| Gender | ||
| Male | 78 | 39 |
| Female | 120 | 61 |
| Age (years) | ||
| Median | 62 | |
| Range | 23–89 | |
| Ethnicity | ||
| Caucasian | 102 | 52 |
| Asian | 49 | 25 |
| Hispanic | 28 | 14 |
| AA | 11 | 6 |
| Other | 8 | 4 |
| Stage | ||
| I | 32 | 16 |
| II | 57 | 29 |
| III | 64 | 32 |
| IV | 45 | 23 |
| Tumor type | ||
| GEC | 42 | 21 |
| Pancreas | 26 | 13 |
| Hepatobiliary | 15 | 7 |
| Small bowel | 5 | 3 |
| Appendix | 13 | 7 |
| Colon | 65 | 34 |
| Rectum | 25 | 13 |
| Anal | 2 | 1 |
| MSI-high | ||
| Yes | 12 | 6 |
| No | 66 | 33 |
| Unknown | 120 | 61 |
| Tumor grade | ||
| Low | 25 | 13 |
| Moderate | 72 | 35 |
| High | 44 | 22 |
| Unknown | 57 | 29 |
| Site of metastasis | ||
| Liver | 24 | 12 |
| Lung | 11 | 6 |
| Peritoneum | 17 | 9 |
| Other | 8 | 4 |
| Treatment modality | ||
| Surgery | 182 | 92 |
| Radiation | 24 | 12 |
| Systemic Treatment | 165 | 83 |
| Regimen | ||
| Fluoropyrimidine based | 136 | 68 |
| Gemcitabine based | 16 | 8 |
| Taxane based | 11 | 6 |
| Biologics | 39 | 20 |
| Serial testing | ||
| Total | 64 | 100 |
| Neg -> Neg | 42 | 66 |
| Pos -> Pos | 9 | 14 |
| Pos -> Neg | 10 | 16 |
| Neg -> Pos | 3 | 5 |
AA, African American; GEC, gastric/esophageal carcinomas; Neg, negative; Pos, positive; due to rounding, percentages might not exactly add up to 100%.
With regards to tumor characteristics, all stages were represented, including 23% advanced/metastatic tumors. Twenty-four percent of the tumors originated in the upper GI tract, 20% were hepato-pancreato-biliary, and the remaining tumors (56%) were located in the lower GI tract. In patients with metastases, the predominant site of distant metastases was liver (40%), followed by peritoneum and lung (28.3% and 18.3%, respectively).
The majority of patients (92%) had undergone surgical resection. Additionally, 83% had received systemic treatment. The most common chemotherapy regimens were fluoropyrimidine based (68%), while 20% of the patients received checkpoint inhibitors or targeted therapies.
ctDNA status based on patient and tumor characteristics
We next sought to determine whether there might be an association between the available patient/tumor characteristics and the rate of ctDNA positivity. For this purpose, all available plasma samples were included in the analysis. As shown in Table 2, 67 of 258 unique samples were ctDNA positive (26%). There appeared to be a statistically significant correlation between disease stage, number and site of metastases with the rate of ctDNA positivity. Whereas only about 20% of postoperative samples from locoregional tumors (i.e., stage I-III) were ctDNA positive, the rate in stage IV GI cancers was 44% (P=0.004). Plasma samples from patient with distant metastases had a higher median ctDNA positivity rate (36 of 85 samples, 42%) than samples from non-metastatic tumors (31 of 173 samples, 22%) (P<0.00003). Interestingly, while for most sites of metastases the positivity rate ranged between 37–53% (Table 2), 8 of 11 (73%) of the samples from lung metastases were positive. Finally, samples from patients with two or more metastatic sites had a more than double positivity rate compared to samples from patients with only one site of metastasis (19 of 29, 66% vs. 17 of 56, 30%) (P<0.002).
Table 2
| ctDNA | P value | |||||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| N | % | N | % | |||
| Total | 67 | 26 | 191 | 74 | ||
| Gender | ||||||
| Male | 46 | 30 | 105 | 70 | 0.0505 | |
| Female | 21 | 20 | 86 | 80 | ||
| Stage | ||||||
| I | 6 | 21 | 23 | 79 | 0.00356 | |
| II | 11 | 18 | 50 | 82 | ||
| III | 14 | 20 | 55 | 80 | ||
| IV | 26 | 44 | 33 | 56 | ||
| Ethnicity | ||||||
| Caucasian | 35 | 25 | 106 | 75 | 0.14 | |
| Asian | 16 | 30 | 38 | 70 | ||
| Hispanic | 6 | 19 | 26 | 81 | ||
| AA | 8 | 44 | 10 | 56 | ||
| Other | 0 | 0 | 12 | 100 | ||
| Tumor grade | ||||||
| Low | 4 | 17 | 20 | 83 | 0.17 | |
| Moderate | 24 | 28 | 63 | 72 | ||
| High | 19 | 37 | 32 | 63 | ||
| Tumor type | ||||||
| GEC | 7 | 17 | 34 | 83 | 0.3 | |
| Pancreas | 11 | 30 | 26 | 70 | ||
| Hepatobiliary | 6 | 32 | 13 | 68 | ||
| Small bowel | 1 | 17 | 5 | 83 | ||
| Appendix | 4 | 25 | 12 | 75 | ||
| Colon | 27 | 26 | 77 | 74 | ||
| Rectum | 9 | 27 | 24 | 73 | ||
| Anal | 2 | 100 | 0 | 0 | ||
| MSI-high | ||||||
| Yes | 5 | 42 | 7 | 58 | 0.45 | |
| No | 22 | 31 | 50 | 69 | ||
| Systemic treatment | ||||||
| Yes | 24 | 31 | 53 | 69 | 0.09 | |
| No | 36 | 21 | 134 | 79 | ||
| Site of metastasis | ||||||
| Liver | 10 | 37 | 17 | 63 | 0.04 | |
| Lung | 8 | 73 | 3 | 27 | ||
| Peritoneum | 10 | 53 | 9 | 47 | ||
| Other | 7 | 26 | 20 | 74 | ||
| No. of metastatic sites | ||||||
| 0 | 31 | 22 | 142 | 78 | <0.0001 | |
| 1 | 17 | 30 | 39 | 70 | ||
| 2+ | 19 | 66 | 10 | 34 | ||
AA, African American; GEC, gastric/esophageal carcinomas; due to rounding, percentages might not exactly add up to 100%.
Finally, we observed trends for higher ctDNA positivity rate (defined as 10% or higher absolute difference in positivity rate) with male gender (30% vs. 20%), higher tumor grade (37% high grade vs. 17% low grade), tumor location (17% upper GI tract vs. 28% other) and having systemic treatment (31% vs. 21%), although none reached the level of statistical significance.
Despite an average positivity rate of 26%, discrepancies were seen between different ethnicities, with positivity rates ranging from 19% (Hispanic) to 44% (African American). Given the correlation with advanced stage and ctDNA positivity rate, we first determined the race/ethnic distribution among Stage IV patients; Caucasian (26/102, 25.5%), Asian (4/49, 8.2%), Hispanic (4/24, 14.3%), and African American (4/11, 36.4%). Among the patients with stage IV disease, there appeared to be a wide range, with Hispanics showing a trend for the lowest positivity rate (1/6, 16.7%), followed by Caucasian (17/43, 39.5%) and Asian (6/11, 54.5%) patients, and the highest observed rate seen in African American patients (4/5, 80%). This observed effect was less pronounced in Stage I-III patients, although the trends persisted: Hispanic 19.2%, Caucasian 19.4%, Asian 25.6%, and African American 30.8% (Table 3).
Table 3
| Ethnicity | Stage | No. Met. Sites | ctDNA | ||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||
| N | % | N | % | ||||
| Caucasian | I-III | 0 | 19 | 19 | 79 | 81 | |
| IV | 1 | 12 | 36 | 21 | 64 | ||
| >1 | 5 | 50 | 5 | 50 | |||
| Asian | I-III | 0 | 11 | 26 | 32 | 74 | |
| IV | 1 | 5 | 56 | 4 | 44 | ||
| >1 | 1 | 50 | 1 | 50 | |||
| Hispanic | I-III | 0 | 5 | 19 | 21 | 81 | |
| IV | 1 | 0 | 0 | 5 | 100 | ||
| >1 | 1 | 100 | 0 | 0 | |||
| AA | I-III | 0 | 4 | 31 | 9 | 69 | |
| IV | 1 | 0 | 0 | 1 | 100 | ||
| >1 | 4 | 100 | 0 | 0 | |||
No. Met. Sites, number of metastatic sites; AA, African American.
Serial measurement of ctDNA
A total of 64 patients had serial testing, i.e., more than one plasma sample timepoint. Two-thirds of them (42 of 64, 66%) exhibited negative ctDNA throughout. In nine patients (14%), the result remained positive, i.e., no change with treatment. Fourteen patients (22%) had ctDNA status that changed in the course of their treatment. Ten patients (16%) had ctDNA that turned from positive to negative after receiving chemotherapy.
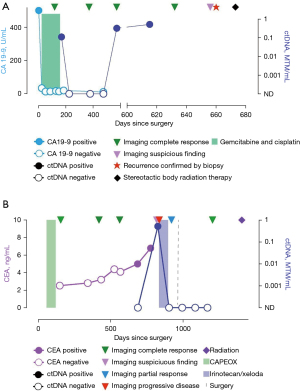
Unfortunately, four patients (6%) had a baseline negative ctDNA that turned positive during the course of their disease. In the first case, the patient had pancreatic adenocarcinoma status post Whipple procedure and adjuvant chemotherapy with 5FU/Leucovorin/oxaliplatin/irinotecan (FOLFIRINOX). ctDNA after surgery and again after six months of adjuvant chemotherapy was negative, however became positive 11.3 months later on surveillance. At that time, the patient was found to have multiple liver metastases on imaging. In the second case, the patient had colon cancer treated with left hemicolectomy and adjuvant capecitabine/oxaliplatin (CAPEOX). ctDNA was negative post completion of adjuvant treatment, then turned positive after 27.5 months in the presence of a single site recurrence close to the ureter, which was identified on CT imaging (Figure 1A). In the third case, the patient had stage IV gastric adenocarcinoma complicated by malignant ascites, and was treated with docetaxel, oxaliplatin, leucovorin, and 5-FU (FLOT) followed by subtotal gastrectomy. 2.9 months later, her ctDNA became positive and a mammogram showed a single focus of metastasis, biopsy consistent with carcinoma of gastric origin with signet ring cells. The patient underwent lumpectomy. The fourth patient was initially diagnosed with Stage II intrahepatic cholangiocarcinoma which was resected and treated with six months of adjuvant chemotherapy. Routine surveillance with CT imaging, CA19-9 remained negative for about a year. Repeat ctDNA testing became positive after 19 months, short-term follow-up a month later confirmed rising ctDNA levels. Ultimately a PET CT showed a solitary T9 vertebral FDG avid lesion, confirmed on MRI of the T spine. The patient underwent treatment for the solitary site of recurrence with stereotactic body surgery (Figure 1B). In all of these cases, ctDNA turned positive with associated disease recurrence.
Discussion
Tumor informed ctDNA assessment is associated with higher sensitivity, compared to tumor agnostic, or ‘static’, ctDNA assays. Hence, it can be used to detect very low disease volumes, i.e., MRD and early recurrence, but also due to its high specificity may serve as a valuable treatment monitoring tool for advanced disease (13).
In GI malignancies, the main body of data for MRD testing has been primarily produced for colon cancer (11-13). While there are emerging data for other GI malignancies (14,15), there is a need for more real-life data for the utility of the MRD assay in other GI and hepatobiliary malignancies. In addition, there has been an underrepresentation of ethnic minorities in currently available data sets. In this work, we describe the feasibility and clinical utility of the ctDNA MRD assay in one of the largest cohorts presented to date. Importantly, two-thirds of the tumors were of non-colon cancer origin, and more than 40% from patients from ethnic minorities. While according to the most current Census data African Americans, Hispanics and Asians comprise about 13.4%, 18.5% and 5.9% of the U.S. population, respectively, most published series to date include 80% or higher Caucasian patients (11,12). Our cohort had a much higher representation of these groups (combined 49%). The majority of the patients in our study were treated at the University of California Irvine located in Orange County, CA, USA. The population of Orange County includes about 55% Hispanics and Asians (16). On the other hand, the distribution by ethnicity in Kansas City, MO, location of the second study site, are 54.9% Caucasian, 28% African American and 6.2% Hispanic. Hence likely the over representation of ethnic minorities in our series.
It has been suggested that in many subsets of GI cancers, African Americans tend to have delayed diagnosis and present with more advanced disease due to socio-economic disparities and lack of access to healthcare (17). In colorectal cancer, African American patients tend to have higher frequencies of KRAS mutations, which might lead to more aggressive cancers (18).
We confirm an association of ctDNA positivity rate with stage of disease (13), whereby the most significant difference is seen in Stage IV tumors compared to non-metastatic disease. While we observed roughly a doubling of positivity rate with each additional site of metastases (i.e., from non-metastatic to single site of metastasis to two or more sites), it is notable that even in Stage I and II disease, there was a 21% and 18% rate of positive ctDNA. These rates are much higher than the 8% positivity rate reported in the study by Tie et al., in which single tumor-specific mutation was used to detect post-operative ctDNA in stage II colon cancer (19). The observed difference could mainly be attributed to the increased sensitivity associated with the use of multiple tumor-specific mutations (8). Considering the very high positive predictive value of the ctDNA MRD assay (11,12), this finding highlights the importance of determining the microscopic ‘post-operative stage’ vs. the traditional anatomic staging which is probabilistic in nature but does not describe the presence of MRD in individual patients, independent of their anatomic stage. Longer follow-up of these patients will provide further clarification regarding rates and patterns of recurrence in Stage I GI cancers with a positive ctDNA MRD test.
We also observed varying rates of ctDNA positivity based on metastatic site. As expected for GI malignancies, the most common site of metastasis was the liver (12% of all patients). However, we could not confirm previous reports of liver metastases being associated with higher ctDNA rates (20,21). On the other hand, we observed a 73% positivity rate for lung metastases, which might possibly indicate more disseminated diseases (since we only included GI cancers), or biologically explained by the abundance of pulmonary capillary bed. Interestingly, previous work has identified peritoneal carcinomatosis with reduced levels of detectable ctDNA (20-22). In our series, 10/19 samples (53%) from patients with peritoneal carcinomatosis had positive ctDNA, highlighting the potential benefit of using a tumor informed assay to increase sensitivity for ctDNA detection in this group of patients. Of note, all patients with peritoneal metastases did not have any other documented site of metastatic spread. Finally, we were able to confirm that primary tumor location of colorectal carcinomas does not appear to affect the rate of ctDNA MRD positivity (23).
Thirteen patients in this study had ctDNA that changed from positive to negative or vice versa. Ten of these patients previously had positive ctDNA, that became negative during or after receiving chemotherapy, suggesting that ctDNA can be used to monitor treatment and remission of disease. Our results also show that patients receiving active systemic treatment have higher rates of positive ctDNA (31%) than those not receiving active chemotherapy (21%). This may reflect that ctDNA positivity rates are higher during active disease when patients are receiving treatment due to higher tumor burden, compared to patients who are not receiving therapy. Another explanation for this observation would be a transient flare of detectable ctDNA due to cell kill and release of ctDNA from systemic treatment.
The retrospective nature of this study is certainly a limitation with regards to accuracy and completeness of all relevant clinical and pathologic characteristics. Additionally, the personalized ctDNA-MRD ctDNA assay only became available outside of clinical trials in late 2019, so the follow-up time for patients with early-stage disease is still too short to fully capture the risk and pattern of recurrence. Longer follow-up, and additional serial testing, will surely address this issue. The relatively small sample size in sub-groups of patients likely affected statistical significance. However, the total number of patients of almost 200 makes this one of the largest reported cohorts, including 49% ethnic minorities, which adds to the available literature and may provide a framework to build upon for further research.
In summary, we provide further evidence for the real-world clinical feasibility and utility of personalized ctDNA MRD testing using a tumor informed assay in a cohort of patients with a wide range of GI malignancies, stages, and ethnic backgrounds.
Acknowledgments
The authors would like to acknowledge the support of Nicole Hook and Shruti Sharma from Natera, Inc. for performing data abstraction and analysis on the patient-specific plots.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-484
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-484
Peer Review File: Available at https://dx.doi.org/10.21037/jgo-21-484
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-484). FD receives research support to institution from Astrazeneca, BMS, Merck, Exelixis, Taiho, Trishula, Signatera, Amgen, Genentech/Roche, Ipsen; honoraria from Exelixis, Deciphera, Sirtex Medical, Amgen, Eisai, Natera Inc., QED; and consulting fees from Exelixis, Foundation One, Genentech/Roche, Eisai, Ipsen, Astrazeneca. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board at the University of California Irvine (protocol# 2020-6196) and Saint Luke’s Health System (protocol# 001CTDNA21) and individual consent for this retrospective anonymized study was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Khorana AA, McKernin SE, Berlin J, et al. Potentially Curable Pancreatic Adenocarcinoma: ASCO Clinical Practice Guideline Update. J Clin Oncol 2019;37:2082-8. [Crossref] [PubMed]
- Shah MA, Kennedy EB, Catenacci DV, et al. Treatment of Locally Advanced Esophageal Carcinoma: ASCO Guideline. J Clin Oncol 2020;38:2677-94. [Crossref] [PubMed]
- Lieu C, Kennedy EB, Bergsland E, et al. Duration of Oxaliplatin-Containing Adjuvant Therapy for Stage III Colon Cancer: ASCO Clinical Practice Guideline. J Clin Oncol 2019;37:1436-47. [Crossref] [PubMed]
- Holdenrieder S, Pagliaro L, Morgenstern D, et al. Clinically Meaningful Use of Blood Tumor Markers in Oncology. Biomed Res Int 2016;2016:9795269. [Crossref] [PubMed]
- Bello Roufai D, Bidard FC. Impact of circulating tumor DNA early detection and serial monitoring in the management of stage I to III colorectal cancer. Ann Transl Med 2019;7:S315. [Crossref] [PubMed]
- Benson AB, Venook AP, Al-Hawary MM, et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;19:329-59. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Coombes RC, Page K, Salari R, et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin Cancer Res 2019;25:4255-63. [Crossref] [PubMed]
- Liu J, Chen X, Wang J, et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann Oncol 2019;30:464-70. [Crossref] [PubMed]
- Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019;5:1124-31. [Crossref] [PubMed]
- Tarazona N, Henriksen TV, Carbonell-Asins JA, et al. Circulating tumor DNA to detect minimal residual disease, response to adjuvant therapy, and identify patients at high risk of recurrence in patients with stage I-III CRC. J Clin Oncol 2020;38:4009. [Crossref]
- Kasi PM, Dayyani F, Morris VK, et al. Tumor-informed assessment of molecular residual disease and its incorporation into practice for patients with early and advanced-stage colorectal cancer (CRC-MRD Consortia). J Clin Oncol 2020;38:4108. [Crossref]
- ctDNA and prognosis in resected esophageal adenocarcinoma (EAC) | OncologyPRO. Available online: https://oncologypro.esmo.org/meeting-resources/esmo-asia-virtual-congress-2020/ctdna-and-prognosis-in-resected-esophageal-adenocarcinoma-eac. Accessed April 4, 2021.
- Hsu CH, Lu S, Abbas A, et al. Longitudinal and personalized detection of circulating tumor DNA (ctDNA) for monitoring efficacy of atezolizumab plus bevacizumab in patients with unresectable hepatocellular carcinoma (HCC). J Clin Oncol 2020;38:3531. [Crossref]
- U.S. Census Bureau QuickFacts: Orange County, California. Available online: https://www.census.gov/quickfacts/orangecountycalifornia. Accessed April 07, 2021.
- Ashktorab H, Kupfer SS, Brim H, et al. Racial Disparity in Gastrointestinal Cancer Risk. Gastroenterology 2017;153:910-23. [Crossref] [PubMed]
- Watson R, Liu TC, Ruzinova MB. High frequency of KRAS mutation in early onset colorectal adenocarcinoma: implications for pathogenesis. Hum Pathol 2016;56:163-70. [Crossref] [PubMed]
- Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [Crossref] [PubMed]
- Osumi H, Shinozaki E, Takeda Y, et al. Clinical relevance of circulating tumor DNA assessed through deep sequencing in patients with metastatic colorectal cancer. Cancer Med 2019;8:408-17. [Crossref] [PubMed]
- Osumi H, Shinozaki E, Yamaguchi K, et al. Clinical utility of circulating tumor DNA for colorectal cancer. Cancer Sci 2019;110:1148-55. [Crossref] [PubMed]
- Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 2017;28:1325-32. [Crossref] [PubMed]
- Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015;26:1715-22. [Crossref] [PubMed]


