
Analysis of dynamic molecular networks: the progression from colorectal adenoma to cancer
Introduction
In the past, due to the disease characteristics of colorectal cancer (CRC), it was rarely diagnosed and treated in a timely manner. Currently, CRC accounts for approximately 10% of annually diagnosed cancers and cancer-related deaths globally. The incidence rate of CRC is second only to breast cancer in women and prostate cancer in men, and it is the fourth most deadly disease worldwide (1,2). Most CRCs originate from colorectal polyps (3). As the use of endoscopes has become more popular, colorectal polyps have been found to be highly prevalent in adults (4). In fact, most polyps are benign, and only a very small number of polyps eventually progress into CRC. The appropriate detection, evaluation, and resection of colorectal polyps can prevent the occurrence and development of CRC. However, the results of existing detection methods are inaccurate, and additional time and professional knowledge in identifying the pathologies of CRC are required. Moreover, the general method for polypectomy is inefficient and increases the total cost. Therefore, it is important to determine the incidence of polyps in CRC for more accurate screening. With the progression of precancerous colorectal adenoma to CRC, whether via a classic adenoma-carcinoma sequence or serrated polyp pathway, defective DNA repair, chromosomal instability, and DNA methylation changes are often used in clinical applications (5,6). However, as polyp size is directly associated with important histological features such as high-grade dysplasia and invasive cancer, it is likely to represent a more clinically effective and cost-effective strategy (7,8).
Polyp size is used as a simple but representative marker to measure the occurrence and development of CRC and, more importantly, to explore its molecular pathogenesis and underlying mechanisms. Dynamic network analysis, a method that has emerged in recent years, can analyze and infer the multifactorial changes of cancer at different stages and recreate a dynamic monitoring network to establish connections (9). Dynamic network analysis is a useful method for understanding the potential mechanisms of cancer development and has been used to investigate the occurrence and development of cancers, including pancreatic ductal carcinoma and breast cancer, to provide references for diagnosing disease progression (10). A recently reported article on the progression of colorectal cancer used transcriptome sequencing and dynamic networks to greatly enrich the concept of classic adenoma-carcinoma sequences. The article analyzes the transcriptomic characteristics of primary CRC, adenoma and normal colonic epithelial tissues to determine six specific dynamic expression patterns during tumor formation, and from this screened the poor prognostic marker TPD52L1 for colorectal cancer. Has great enlightenment for this experiment (11).
Driven by time- and background-dependent changes in multiple genes, cancer is a complex process of interaction between genes and internal and external factors and an accumulation of mutations in key genes of somatic cells. By determining the role of gene mutations in this process, cancer occurrence can be curbed through timely blocking of the change process. Dynamic network analysis can be used to integrate information concerning the genetic changes of polyps at different stages, providing useful data for disease management. This study uses the new Cytoscape application DyNetViewer to construct, analyze and visualize the dynamic network, and describe the dynamic development process from colorectal polyps to colorectal cancer. Compared with adenomas with a higher degree of malignancy, this study used 20 mm as the dividing line to provide preliminary results of early polyps and found representative biomarkers.
We present the following article in accordance with the STREGA reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-674).
Methods
Gene chip screening and data merging
The colorectal adenoma (GSE8671) (12) and colorectal cancer (GSE32323) (13) gene chip data collections used in this experiment were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). The 2 chips contained 49 normal samples, 32 colorectal adenoma samples, and 17 CRC samples (Table 1), The grouping reference is based on the European Society of Gastrointestinal Endoscopy (ESGE) guidelines (2020), which proposes that when the size of polyps is less than 20 mm, cohort studies based on the risk of CRC rather than the risk of metachronous advanced tumors usually reduce diversity and impact. When the polyp size is ≥20 mm, it is closely related to the long-term CRC incidence/increased risk of death after polypectomy. Therefore, we use 2cm as the grouping standard (14). The affy (15) package in R language software was used to read the CEL file corresponding to the samples, the robust multichip average algorithm was utilized to process and generate the probe expression profile a, and the “removeBatchEffect” function of the R package limma (16) was used to eliminate potential batch processing effect and to obtain the probe expression profile b (17). The R packages FactoMineR and factoextra were used to perform principal component analysis and visualize the 2-probe expression data of a and b, respectively. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Type | Number |
|---|---|
| Tissue | |
| Normal | 49 |
| Polyp | 32 |
| Cancer | 17 |
| Polyp size | |
| ≤2 cm | 23 |
| >2 cm | 9 |
| Cancer stage | |
| Stage I | 4 |
| Stage IIa | 6 |
| Stage IIb | 8 |
| Stage IIIb | 6 |
| Stage IIIc | 4 |
| Stage IV | 6 |
| Location | |
| Transversum colon | 2 |
| Ascending colon | 8 |
| Descending colon | 8 |
| Sigmoid colon | 32 |
| Rectum | 14 |
| NA | 17 |
Screening and processing of differential genes
The samples in the 2 chips were divided into 4 groups representing the 4 stages of the progression from colorectal adenoma to CRC: normal, polyp size ≤2 cm, polyp size >2 cm, and tumor. We then used the limma package and c file in R to screen the 3 groups for genes that differed from the normal group. The critical standard |log2 fold change (FC)| was >2, and false discovery rate (FDR) was <0.01. The UpSetR package in R was then used to intersect the up- and downregulated differential genes. The 3 sets of differential genes were combined, and the gene expression of the union genes corresponding to the samples in the 4 groups was averaged. The gene expression matrix was further filtered to include the average difference of the top 75% most variable genes to obtain the combined differential gene data matrix of expression changes in the development of CRC.
Time course protein interaction network processing
The union differential gene in the previous step was retrieved from the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org), and the corresponding static protein-protein interaction (PPI) network tab-separated value (TSV) file was exported. The TSV file was imported into Cytoscape 3.6.1 software for visualization. The DyNetViewer (18) plugin for visualizing dynamic networks in Cytoscape 3.0 was used to construct the time course protein interaction network (TC-PIN). The TC-PIN algorithm constructed a subnetwork, and the threshold was set to the quartile point of the data in the just change data matrix: 5.6959891525. In the dynamic node analysis option, all nodes were selected, and all node attribute change values were output. Seventy-seven dynamic network characteristic genes were then screened out for each stage of the entire disease development process and attribute change diagrams were drawn.
The time-sequenced network-based protein complex discovery algorithm and modularity attribute was selected to analyze dynamic network clusters. A total of 25 network modules were obtained, of which 19 modules were characteristic of various stages.
Feature network module enrichment analysis
The R package Pi was used to perform a weighted ranking of the average expression of each node at each stage based on the random walk with restart algorithm. Subsequently, the xPierGSEA function was used to perform gene analysis in combination with ranking gene set enrichment analysis. The parameter setting random number seed was 1,234, and the number of iterations was 5,000. The enrichment background was set to “MsigdbC5BP” and “MsigdbC2KEGG” from the Molecular Signatures Database (MSigDB), and the correlation with CRC was obtained. The distribution trend in the gene table helped to more accurately identify protein complexes.
Weighted Gene Co-Expression Network Analysis (WGCNA)
Use the pickSoftThreshold (19) function to construct a scale-free co-expression network with the expression data profiles of the 5,047 genes that were screened. Then, network construction and module detection are performed by the one-step function “blockwiseModules” (20). The R function of blockwiseModules has many parameters; in our research, the parameters are implemented as follows: power =8, maxBlockSize =5047, minModuleSize =30 and networkType = “unsigned”.
Statistical analysis
The statistical analysis of DEG is done in R software. The cumulative survival time was calculated by Kaplan-Meier method and analyzed by log-rank test. Pearson χ2 test and Student’s t test are used for comparison between groups. Probability (P) values less than 0.05 are considered statistically significant (*P<0.05; **P<0.01; **P<0.001).
Results
Combining gene chip data sets to eliminate batch effects
The colorectal adenoma (GSE8671) and colorectal cancer (GSE32323) gene chip data collections were obtained from the GEO database. Considering the different backgrounds of the 2 chips, there was a strong batch effect, which was also confirmed when the principal component analysis chart was generated (Figure 1A); thus, we removed the batch effect and obtained a more accurate analysis chart (Figure 1A,1B).
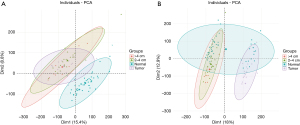
Identification and expression pattern analysis of differentially expressed genes at different stages of colon cancer
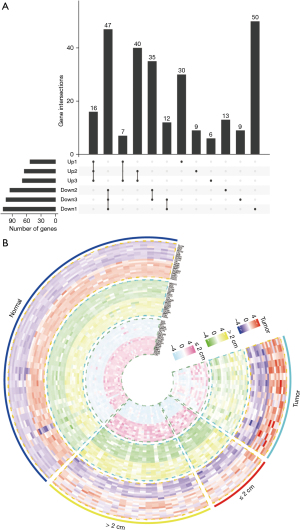
The differentially expressed genes (DEGs) that were significantly dysregulated at different stages of CRC (normal, polyp size ≤2 cm, polyp size >2 cm, tumor) were screened out based on R package analysis. Compared with normal tissue, polyp size ≤2 cm, polyp size >2 cm, and the DEGs that were significantly upregulated and downregulated at each stage were selected as the intersection of different stages (Figure 2A). The top 10 upregulated and downregulated DEGs were clustered and displayed in a hierarchical heat map at each stage (Figure 2B). The 20 highest DEGs could clearly distinguish the various stages of CRC in normal colorectal tissue. Changes in related genes were observed.
Construction of PPI network
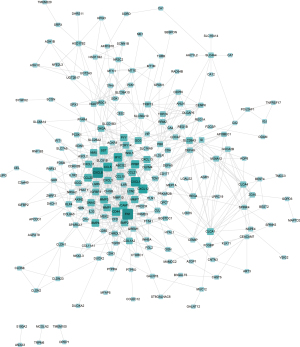
The online database STRING was used to construct a PPI network for CRC and DEGs of normal colorectal tissue at different stages. The union of each DEG established a PPI network. As shown in Figure 3, the network consisted of 214 nodes interacting with 746 edges.
Data clustering analysis of dynamic network
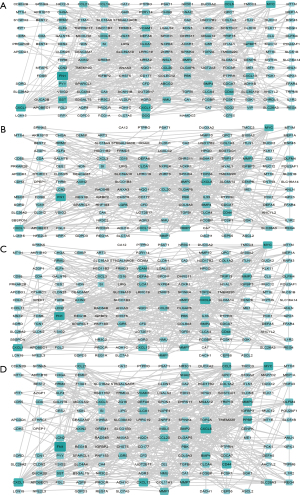
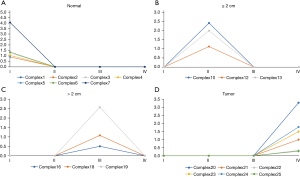
DyNetViewer is a Cytoscape application for dynamic network construction, analysis, and visualization. To analyze the dynamic clustering properties of progressing CRC, we input the 4-stage network into DyNetViewer for visualization and used the construct dynamic network function of the DyNetViewer plugin to construct a dynamic protein interaction network. The TC-PIN algorithm constructed subnetworks, showing clusters of highly interconnected regions at each stage, and these subnetworks were analyzed as key regulators of network development (Figure 4). A total of 25 feature cluster modules were identified in the 4 CRC stages. In the first stage, 7 groups were typical contributions to the network. These 7 gene sets were involved in: alcohol metabolism; amino acid metabolism; cell cycle regulation; blood pressure and electrolyte homeostasis; protein processing and assembly; chemotactic activity of inflammatory cells; intestinal contraction; blood sugar balance; vasodilation, and; hematopoietic coagulation. In addition, we found that the key genes ADH1C, ADH1B, and UGT2A3 were involved in up to 4 pathways, respectively, in the first stage (Figure 5A). We found that the second stage of the network involved 3 clusters. Pathway enrichment showed that these clusters were mainly enriched in: intestinal contraction; fibronectin activity; cartilage and bone formation; cell adhesion and interaction; angiogenesis; collagen production, and; stem cell differentiation. MYC, AXIN2, and SPARCL1 had 4 or more pathways (Figure 5B). In the third stage of the network, 3 gene sets were identified as the key modules of the protein interaction network (Figure 5C). These clusters were related to: intestinal contraction; Helicobacter pylori infection; fibronectin activity; cartilage and bone formation; cell adhesion and interaction; angiogenesis; collagen production, and; stem cell differentiation. Two key genes, CXCL8 and CXCL11, and SPARCL1 were shared by up to 4 pathways. Finally, a total of 6 clusters participated in the fourth stage of the network (Figure 5D). These clusters were mainly enriched in: cell canceration; intestinal contraction; fibronectin activity; cartilage and bone formation; cell adhesion and interaction; angiogenesis; vasoconstriction; collagen production, and; stem cell differentiation. Two key genes, CXCL8 and CXCL1, CCL5, and COL11A1 were shared by up to 4 pathways.
Analysis of node centrality of dynamic network
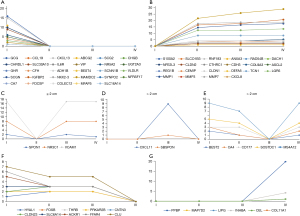
To screen out key genes involved in the dynamic network, dynamic node analysis in the DyNetViewer plugin was used, and the key nodes of each CRC stage were calculated using a degree centrality (DC) measurement. In our research, we identified 77 key genes that might be involved in the development of dynamic networks (Figure 6A-6G). Five genes were specifically found in the normal stage, including CXCL8, MMP7, MMP3, MMP1, and LGR5, with the highest DC. VCAM1 and NR3C1 were the characteristic nodes for a polyp size ≤2 cm. The characteristic nodes of the third stage included SBSPON, CXCL11, and CXCL11, which was the key to maintaining network stability during this stage. At the tumor stage, PPBP, CCL5, SNAI2, CCL21, and SST were the characteristic nodes, and PPBP was the key gene for this node. Five nodes, including CXCL8, MMP3, LGR5, MMP7, and MMP1, were potentially key genes that promoted the occurrence of CRC.
Pathway enrichment of DEGs at different stages
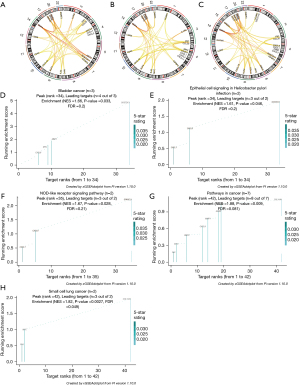
We then predicted the similarity of the gene function of the feature module at each stage to support the possibility of functional level as a module (Figure 7A-7C). To better analyze the module function at each stage and prove our modularity, we used Pi package analysis. Subsequently, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that the upregulation of DEGs at different CRC stages was usually concentrated in 4 key pathways. However, at different stages, each had its own unique characteristic pathway, which was the focus of our attention (Figure 7D-7H).
Dynamic expression patterns associated with pathological phenotype in the adenoma-carcinoma sequence
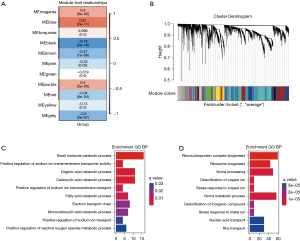
We constructed an adjacency matrix and constructed a topological overlap matrix. Finally, 11 modules were determined based on average hierarchical clustering and mergeCutHeight =0.25 (Figure 8A). The blue and black modules are highly related to pathological grading; therefore, this module was selected as a clinically important module for further analysis (Figure 8B-8D).
Discussion
Current research has shown that most cases of CRC are sporadic, often originating from polyps in abnormal crypts, and only about 10% of such polyps will develop into early adenomas, then advanced adenomas, and finally CRC. This progression is broadly classified as the traditional tubular adenoma pathway or the serrated polyp pathway. Later, because Fearon and Vogelstein proposed their multiple genetic models of colorectal cancer (21), the gradually accumulated insights such as chromosomal instability (CIN), microsatellite instability (MSI) and methylation phenotype have made us think about the role of glands. A deeper understanding of the multiple genetic and epigenetic changes underlying tumor-carcinoma sequence occurrence and development has provided some information about its pathogenesis. The emergence and development of omics technology has further revealed off the characteristics of gene expression at the molecular level (22,23). Earlier and more practicable screening can provide a reliable preliminary basis for the prevention and treatment of CRC. The dynamic molecular mechanism of CRC progression from polyps is still unclear; therefore, in this study, we analyzed microarrays from the GEO database and identified the DEGs of normal tissue, polyps of different sizes, and CRCs. We then used the plugin DyNetViewer to construct a dynamic molecular interaction network and determined functional modules and characteristic nodes of each stage, that is, genes that might be related to CRC progression.
The genetic changes in CRC at different stages are dynamic and complex. Dynamic network analysis described the differential gene changes at various stages, from normal tissue to polyps to cancer. During these stages, MMP1, MMP3, and MMP7, which are all matrix metalloproteinases (MMPs), increased steadily. MMPs are zinc-dependent endopeptidases. The family consists of 25 members (including collagenase, gelatinase, and matrix metalloproteinases) and plays a role in cancer invasion and metastasis. A key role of MMP is being involved in the remodeling of extracellular matrix, which is strongly associated with poor prognosis in cancer (24-26). Previous studies on CRC have also pointed out that the content of MMPs in CRC tissue is significantly upregulated, which is related to metastasis and proliferation in CRC (27,28). In our study, we observed that MMP1, MMP3, and MMP7 were upregulated to the greatest extent during the initial stage, from normal tissue to polyp size ≤2 cm, and also increased slightly in the later stages. Related literature has reported that MMP3 promoted the development of serrated polyps, although this requires further research. Several studies have observed that MMPs were potential biomarkers (29-31).
Similarly, CXCL8 and CXCL7 (PPBP) changed from the initial stage to the tumor stage. However, the increase in CXCL8 was observed during the period from normal tissue to polyp size ≤2 cm, while the increase in CXCL7 was observed at polyp size >2 cm. Several previous studies have proposed the use of these two genes as potential cancer biomarkers at the tumor stage, although they were not subdivided more specifically. Differences between the two genes could be further subdivided to provide a reference for more accurate screening (32-34). The chemokines CCL19 and CXCL13 exhibited results opposite to CXCL8 and CXCL7, with both showing a rapid decline in cancerous tissue. Further results supporting CCL19’s anti-CRC angiogenesis have also been reported (35,36). However, studies reporting on CXCL13 had differing results, which is worthy of discussion (37,38). Finally, the expression of GCG in CRC tissue was significantly downregulated, and GCG is a preproprotein that can be cleaved into 4 different mature peptides (39,40). One of them, glucagon, is a ligand for specific G protein-linked receptors, and its signaling pathway controls cell proliferation. Therefore, we inferred that GCG might have played a major role in inhibiting the occurrence of CRC.
KEGG pathway enrichment analysis showed that the DEGs of polyps and cancer were mainly enriched in 22 pathways, including the bladder cancer pathway, epithelial cell signaling in Helicobacter pylori infection, NOD-like receptor signaling pathway, small cell lung cancer pathway, and CRC pathway (Figure 7D-7H). The interaction pathways between factors and cytokine receptors play an important role in the maintenance of the pathological environment of CRC. These pathways share CXCL8, CXCL1, FN1, MMP1, MYC, and other genes and have been reported to be associated with the occurrence and development of CRC. Distinct pathways were also observed at specific stages. For example, when polyp size >2 cm, leukocyte migration across the endothelium and the appearance of Toll-like receptor signaling pathways indicated that inflammation was an important hallmark in the early stage of CRC (41,42). The few reports on endothelial migration and CRC in leukocytes have found that they serve as a signaling pathway for inflammation and immunomodulation. Immunomodulation has been observed when polyps are >2 cm, and they are high-risk factors for CRC. During the tumor period, cell adhesion molecules (CAMs), which participate in the proliferation and migration of cancer cells, are observed (43). Genes such as VCAM1 and COL11A1 are involved in proliferation and migration. As a classic cancer pathway, their appearance is a high-risk indicator used for screening, while several related therapies are also emerging (44-46). In addition, the mitogen-activated protein kinase signaling pathway and hematopoietic cell lineage are not observed at the cancer stage, which might be associated with abnormal angiogenesis in cancer (47-49).
In this study, we focused on dividing the various stages from polyps to cancer and described the DEGs of each stage to provide references for more accurate early screening. However, the limited number of samples and lack of clinical samples in our study limit their use. During follow-up, samples from clinical trials should be collected to better support our hypothesis.
Conclusions
We screened the DEGs of different CRC stages and constructed a dynamic molecular interaction network to illustrate the underlying mechanisms of CRC progression. The matrix metalloprotein family and chemokines were identified as key regulatory genes driving CRC progression. In addition, the related nodes and pathways at various stages may be potential mechanisms for promoting the dynamic CRC progression. Our research provides a better understanding of the dynamic pattern of molecular interaction networks during the progression of polyps to CRC and provides potential opportunities for therapeutic interventions.
Acknowledgments
Funding: This study was supported by Zhejiang Provincial Natural Science Foundation of China, Zhejiang Province, People’s Republic of China (LYQ20H310001, LY20H310001), the Medical and Health Research Program of Zhejiang Province (2021KY055, 2022KY069, 2022RC008, 2018KY297), the Project of Administration of Traditional Chinese Medicine of Zhejiang Province of China (2021ZZ001, 2017ZA037), the “10000 Talents Plan” of Zhejiang Province (to P Huang, 2020R52029), the Zhejiang Provincial Program for the Cultivation of 151 Talents (to P Huang), the Zhejiang Provincial Program for the Cultivation of High-Level Innovative Health Talents (to P Huang), the Zhejiang Provincial Program for the Cultivation of High-level Innovative Health Talents (to Y Zhang) and the open topic fund from Key Laboratory of Clinical Cancer Pharmacology and Toxicology Research of Zhejiang Province (2020E10021).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-674
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-674). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Pickhardt PJ, Pooler BD, Kim DH, et al. The Natural History of Colorectal Polyps: Overview of Predictive Static and Dynamic Features. Gastroenterol Clin North Am 2018;47:515-36. [Crossref] [PubMed]
- Pamudurthy V, Lodhia N, Konda VJA. Advances in endoscopy for colorectal polyp detection and classification. Proc (Bayl Univ Med Cent) 2020;33:28-35. [Crossref] [PubMed]
- Grady WM, Pritchard CC. Molecular alterations and biomarkers in colorectal cancer. Toxicol Pathol 2014;42:124-39. [Crossref] [PubMed]
- Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16:713-32. [Crossref] [PubMed]
- Pickhardt PJ, Hain KS, Kim DH, et al. Low rates of cancer or high-grade dysplasia in colorectal polyps collected from computed tomography colonography screening. Clin Gastroenterol Hepatol 2010;8:610-5. [Crossref] [PubMed]
- Pickhardt PJ, Kim DH. Colorectal cancer screening with CT colonography: key concepts regarding polyp prevalence, size, histology, morphology, and natural history. AJR Am J Roentgenol 2009;193:40-6. [Crossref] [PubMed]
- Kyrochristos ID, Ziogas DE, Roukos DH. Dynamic genome and transcriptional network-based biomarkers and drugs: precision in breast cancer therapy. Med Res Rev 2019;39:1205-27. [Crossref] [PubMed]
- Pan Z, Li L, Fang Q, et al. Analysis of dynamic molecular networks for pancreatic ductal adenocarcinoma progression. Cancer Cell Int 2018;18:214. [Crossref] [PubMed]
- Hong Q, Li B, Cai X, et al. Transcriptomic Analyses of the Adenoma-Carcinoma Sequence Identify Hallmarks Associated With the Onset of Colorectal Cancer. Front Oncol 2021;11:704531. [Crossref] [PubMed]
- Khamas A, Ishikawa T, Shimokawa K, et al. Screening for epigenetically masked genes in colorectal cancer Using 5-Aza-2'-deoxycytidine, microarray and gene expression profile. Cancer Genomics Proteomics 2012;9:67-75. [PubMed]
- Sabates-Bellver J, Van der Flier LG, de Palo M, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res 2007;5:1263-75. [Crossref] [PubMed]
- Hassan C, Antonelli G, Dumonceau JM, et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline - Update 2020. Endoscopy 2020;52:687-700. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Harbig J, Sprinkle R, Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res 2005;33:e31. [Crossref] [PubMed]
- Li M, Yang J, Wu FX, et al. DyNetViewer: a Cytoscape app for dynamic network construction, analysis and visualization. Bioinformatics 2018;34:1597-9. [Crossref] [PubMed]
- Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol 2008;4:e1000117. [Crossref] [PubMed]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17.
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759-67. [Crossref] [PubMed]
- Kamal Y, Schmit SL, Hoehn HJ, et al. Transcriptomic Differences between Primary Colorectal Adenocarcinomas and Distant Metastases Reveal Metastatic Colorectal Cancer Subtypes. Cancer Res 2019;79:4227-41. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Shay G, Lynch CC, Fingleton B. Moving targets: Emerging roles for MMPs in cancer progression and metastasis. Matrix Biol 2015;44-46:200-6. [Crossref] [PubMed]
- Chen Z, Liu M, Xie K, et al. Huisheng Oral Solution exerts anti-tumor effects by downregulating tissue factor and inhibiting the expression of metastasis-related factors, CD44, MMP2, and VEGF. Transl Cancer Res 2019;8:2602-12. [Crossref]
- Cabral-Pacheco GA, Garza-Veloz I, Castruita-De la Rosa C, et al. The Roles of Matrix Metalloproteinases and Their Inhibitors in Human Diseases. Int J Mol Sci 2020;21:9739. [Crossref] [PubMed]
- Mochizuki S, Ao T, Sugiura T, et al. Expression and Function of a Disintegrin and Metalloproteinases in Cancer-Associated Fibroblasts of Colorectal Cancer. Digestion 2020;101:18-24. [Crossref] [PubMed]
- Herszényi L, Hritz I, Lakatos G, et al. The behavior of matrix metalloproteinases and their inhibitors in colorectal cancer. Int J Mol Sci 2012;13:13240-63. [Crossref] [PubMed]
- Nagata N, Ohta H, Yamada A, et al. Activities of matrix metalloproteinase-2, matrix metalloproteinase-9, and serine proteases in samples of the colorectal mucosa of Miniature Dachshunds with inflammatory colorectal polyps. Am J Vet Res 2020;81:572-80. [Crossref] [PubMed]
- He Z, Chen L, Chen G, et al. Interleukin 1 beta and Matrix Metallopeptidase 3 Contribute to Development of Epidermal Growth Factor Receptor-Dependent Serrated Polyps in Mouse Cecum. Gastroenterology 2019;157:1572-1583.e8. [Crossref] [PubMed]
- Boruk M, Railwah C, Lora A, et al. Elevated S100A9 expression in chronic rhinosinusitis coincides with elevated MMP production and proliferation in vitro. Sci Rep 2020;10:16350. [Crossref] [PubMed]
- Pączek S, Łukaszewicz-Zając M, Gryko M, et al. CXCL-8 in Preoperative Colorectal Cancer Patients: Significance for Diagnosis and Cancer Progression. Int J Mol Sci 2020;21:2040. [Crossref] [PubMed]
- Desurmont T, Skrypek N, Duhamel A, et al. Overexpression of chemokine receptor CXCR2 and ligand CXCL7 in liver metastases from colon cancer is correlated to shorter disease-free and overall survival. Cancer Sci 2015;106:262-9. [Crossref] [PubMed]
- Li L, Zhang L, Tian Y, et al. Serum Chemokine CXCL7 as a Diagnostic Biomarker for Colorectal Cancer. Front Oncol 2019;9:921. [Crossref] [PubMed]
- Xu Z, Zhu C, Chen C, et al. CCL19 suppresses angiogenesis through promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1alpha/VEGF-A pathway in colorectal cancer. Cell Death Dis 2018;9:974. [Crossref] [PubMed]
- Chen L, Lu D, Sun K, et al. Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Gene 2019;692:119-25. [Crossref] [PubMed]
- Zhang G, Luo X, Zhang W, et al. CXCL-13 Regulates Resistance to 5-Fluorouracil in Colorectal Cancer. Cancer Res Treat 2020;52:622-33. [Crossref] [PubMed]
- Zhao S, Mi Y, Guan B, et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol 2020;13:156. [Crossref] [PubMed]
- Gao X, Yang J. Identification of Genes Related to Clinicopathological Characteristics and Prognosis of Patients with Colorectal Cancer. DNA Cell Biol 2020;39:690-9. [Crossref] [PubMed]
- Zhao ZW, Fan XX, Yang LL, et al. The identification of a common different gene expression signature in patients with colorectal cancer. Math Biosci Eng 2019;16:2942-58. [Crossref] [PubMed]
- Chimen M, Apta BH, Mcgettrick HM. Introduction: T Cell Trafficking in Inflammation and Immunity. Methods Mol Biol 2017;1591:73-84. [Crossref] [PubMed]
- Anthoney N, Foldi I, Hidalgo A. Toll and Toll-like receptor signalling in development. Development 2018;145:dev156018. [Crossref] [PubMed]
- Oh HH, Joo YE. Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intest Res 2020;18:168-83. [Crossref] [PubMed]
- Andersson Y, Inderberg EM, Kvalheim G, et al. Immune stimulatory effect of anti-EpCAM immunotoxin - improved overall survival of metastatic colorectal cancer patients. Acta Oncol 2020;59:404-9. [Crossref] [PubMed]
- Dai H, Wu Z, Jia H, et al. Bispecific CAR-T cells targeting both CD19 and CD22 for therapy of adults with relapsed or refractory B cell acute lymphoblastic leukemia. J Hematol Oncol 2020;13:30. [Crossref] [PubMed]
- Pishvaian M, Morse MA, McDevitt J, et al. Phase 1 Dose Escalation Study of MEDI-565, a Bispecific T-Cell Engager that Targets Human Carcinoembryonic Antigen, in Patients With Advanced Gastrointestinal Adenocarcinomas. Clin Colorectal Cancer 2016;15:345-51. [Crossref] [PubMed]
- Wang L, Deng L, Lin N, et al. Berberine inhibits proliferation and apoptosis of vascular smooth muscle cells induced by mechanical stretch via the PDI/ERS and MAPK pathways. Life Sci 2020;259:118253. [Crossref] [PubMed]
- Fan SJ, Kroeger B, Marie PP, et al. Glutamine deprivation alters the origin and function of cancer cell exosomes. EMBO J 2020;39:e103009. [Crossref] [PubMed]
- Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci 2020;77:1745-70. [Crossref] [PubMed]


