
High expression of MAGE-C1 gene in colorectal cancer is associated with its poor prognosis
Introduction
The melanoma antigen gene (MAGE) family, a member of the cancer-testis antigen (CTA) family (1), has attracted increasing attention from researchers. Due to MAGE protein’s immunogenicity, they can be used as new markers for tumor immunotherapy (2). MAGE family proteins have a certain relationship with the aggressiveness of tumors, including poor clinical prognosis, acceleration of tumor progression, and metastasis (3-5). In recent years, numerous in-depth studies have been performed on the MAGE family proteins in tumor immunotherapy and prognosis although many challenges remain.
Several studies have shown an association of the MAGE family proteins to the prognosis of various tumors, such as non-small cell lung cancer (6), melanoma (3,5), breast cancer (7), and prostate cancer (8), and therefore, they are considered to be markers for poor prognosis of many tumors. However, as tumorigenesis is affected by many factors, such as genetic changes and individual patient differences, linking individual MAGE antigen with poor survival rates in tumors is insufficient evidence to conclusively show that their expression promotes aggressive tumor growth or that they are associated with chemorefractory disease.
Mori et al. first observed the expression of MAGE in colorectal cancer (CRC) (9). Recently, it has been reported that MAGE-A3 (10), MAGE-D4 (11), MAGE-A9 (12) are highly expressed in colorectal cancer, and the former two are associated with poor prognosis of colorectal cancer, but the mechanism underlying the occurrence is unclear. Further studies may be helpful to utilize these proteins as targets for immunotherapy or targeted therapies.
MAGE-C1, also known as CT7 or CT7.1, is a member of MAGE-C subfamily which is clustered on the X-chromosome and the protein which encodes contains a large number of unique short repeats ahead of the MAGE homologous sequence and thus is therefore about 800 amino acids longer than other MAGE proteins. Current studies have focused MAGE-C1 on multiple myeloma to explore its value in malignant cell typing (13), and other studies have found it to be associated with poor prognosis of multiple myeloma (14). Some studies have found its abnormal expression in ovarian cancer (15), others in breast cancer and associated with poor prognosis in breast cancer patients (16). However, few studies have reported its expression in colorectal cancer.
In our study, we aimed to establish a model that utilize expression of MAGE-C1, clinicopathological characteristics and genes with clinical concern of CRC to predict the prognosis of CRC. First, the genes with significant expression differences in CRC were analyzed by bioinformatics methods through The Cancer Genome Atlas (TCGA) database. MAGE-C1, an abnormally expressed protein, was identified for its significant expression differences. Then, we investigated the expression of MAGE-C1 in CRC using immunohistochemistry (IHC) and analyzed its relationship with the clinicopathological characteristics of CRC and the related hotspot gene changes. At last, these 3 data sets were adopted to establish a model to comprehensively evaluate the combined effect of MAGE-C1 and clinicopathological characteristics and hotspot gene changes on the prognosis of CRC patients.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-739).
Methods
Bioinformatics analysis
The “edgeR” and “limma” packages in R software (version3.6.2, https://www.r-project.org; The R Foundation for Statistical Computing, Vienna, Austria) were used to evaluate differentially expressed genes (DEGs) of CRC in TCGA (https://portal.gdc.cancer.gov/). The gene expression profiles of CRC were downloaded from TCGA. RNA sequencing (RNA-seq) count data on CRC and corresponding clinical information were freely downloaded by R package GDCRNATools (17). There were 408 CRC samples, including 376 tumor and 32 normal tissues. The DeSeq2 package (18) was used to identify the DEGs genes in CRC. DEGs were defined as genes with P value <0.05 and |fold change (FC)| ≥2. The DEGs were also clustered using the package “Heatmap” and were visualized as Volcano Plots using the “ggplot2” package.
Collection of sample cases with CRC
This study collected 156 paraffin-embedded specimens from the Affiliated Hospital of Jiangnan University, Jiangsu, China, from March 2014 to January 2015. There were 85 males and 71 females, aged 31–89 years, with a median age of 70 years. The clinicopathological data of the above cases were summarized. All the above cases were treated in accordance with Chinese CRC diagnosis and treatment protocols and standards. The data of Kirsten rat sarcoma (KRAS), v-RAF murine sarcoma viral oncogene homolog B1 (BRAF), and human epidermal growth factor receptor 2 (HER2) genes were procured from routine testing items in the pathology department of the hospital. The follow-up time was considered to be from surgical removal of the patient’s tumor tissue to the patient’s tumor-related death. The collection and processing of specimens were performed after informed consent of the patients was obtained. The study was approved by the Ethics Committee of the Affiliated Hospital of Jiangnan University (No. 2014-012-001). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
IHC analysis
MAGE-C1 primary antibody (rabbit monoclonal: EPR18067) was purchased from Abcam (Cambridge, UK), and DAB color reagent was purchased from Gene Technology Shanghai Co., Ltd, (Shanghai, China). The specimens were immediately immersed in 10% neutral formalin fixative and fixed from 4 to 16 hours after collection. After collection by the pathologist, the specimens were dehydrated, embedded, and then cut into sections, each 4 μm in thickness. The sections were then deparaffinized with xylene, dehydrated with gradient ethanol, immersed in citric acid sodium citrate buffer for antigen repair, blocked with 3% normal goat serum, and incubated with the primary antibody for 1 hour and the secondary antibody for 30 minutes. Finally, they were stained with DAB. The IHC results were interpreted by 2 pathologists. In this study, a semiquantitative method was used to evaluate the results of IHC. The brown color of the cell membrane and/or cytoplasm indicated positive MAGE-C1 protein IHC. The intensity of staining was divided into 4 levels: no staining (−), 0 points; weak positive (+), 1 point; medium intensity (++), 2 points; and strong positive (+++), 3 points. The density of MAGE-C1-positive cells was divided into 4 levels: no staining, 0 points; proportion of positive cells >0 and ≤10%, 0.1 points; proportion of positive cells >10% and ≤50%, 0.5 points; and proportion of positive cells greater than 50%, 1 point.
For semiquantitative scoring, an H score was used. The relative formula was the following: H = staining intensity × staining density. The cutoff value was set at 1.0 according to the clinical testing standard of the pathology department. When the value was above and equal to the cutoff value, this indicated high expression; when the value was lower than the cutoff value, this indicated low expression.
Statistical analysis
All statistical analyses in this study were carried out using R statistical software. A chi-square test was used to assess the correlation between MAGE-C1 expression and the clinical characteristics of the patients. The effect of MAGE-C1 expression on the overall survival rate was analyzed by the Kaplan-Meier method, and the difference was ascertained via a log-rank test. The risk factors used in prognoses for CRC patients were analyzed by Cox regression models. A nomogram was constructed based on the results of the multivariate analysis using the “rms” package in R version 3.6.2. The reported statistical significance levels were all two sided, with the statistical significance set at a P value <0.05.
Results
Identification of MAGE-C1 in CRC by bioinformatics analysis
The messenger RNA (mRNA) data of CRC tissue were downloaded from TCGA database, and totally 18,473 genes were included for analysis by DESeq2 after merging of the same genes. We found that 2,806 genes were upregulated in CRC tissues and 2,618 genes were downregulated (Figure 1: heat map, volcano plot and violin plot) after threshold value analysis (P<0.01, |fold change| >2). We observed that MAGE-C1 (LogFC =8.87; false discovery rate =1.99×10−7) was one of the most significant outlier genes in the list.
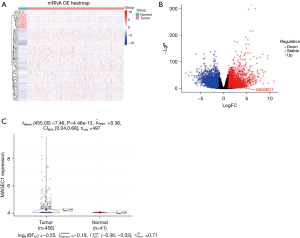
The MAGE-C1 expression in CRC and its relationship with clinicopathological characteristics, CRC hotspot gene mutations, and prognosis
We detected MAGE-C1 expression in paraffin-embedded tissues of 156 patients with CRC. IHC results showed that MAGE-C1 was mainly expressed in the cell membrane and cytoplasm, as shown in Figure 2; MAGE-C1 was not expressed in normal colorectal mucosa controls. The proportions of high and low MAGE-C1 expression in CRC were 28.8% (45/156) and 71.2% (111/156), respectively. The MAGE-C1 expression in CRC was related to the cancer T stage (P=0.010) and M stage (P<0.001) of patients, which means tumor invasion, and metastasis were related to the expression of MAGE-C1, as shown in Table 1.
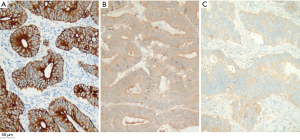
Table 1
| Characteristics | No. | High | Low | P value |
|---|---|---|---|---|
| Age, years | 0.993 | |||
| ≤60 | 41 | 12 | 29 | |
| >60 | 115 | 33 | 82 | |
| Gender | 0.866 | |||
| Female | 71 | 20 | 51 | |
| Male | 85 | 25 | 60 | |
| T stage | 0.010 | |||
| T1–2 | 37 | 4 | 33 | |
| T3–4 | 119 | 41 | 78 | |
| N stage | 0.426 | |||
| N0 | 96 | 25 | 71 | |
| N1–N2 | 60 | 20 | 40 | |
| M stage | 0.001 | |||
| M0 | 124 | 27 | 97 | |
| M1 | 32 | 18 | 14 | |
| Differentiation | 0.309 | |||
| Poorly | 39 | 14 | 25 | |
| Well/moderately | 117 | 31 | 86 | |
| HER2 | 0.984 | |||
| Wild | 152 | 44 | 108 | |
| Mutation | 4 | 1 | 3 | |
| KRAS | 0.086 | |||
| Wild | 101 | 24 | 77 | |
| Mutation | 55 | 21 | 34 | |
| BRAF | 0.424 | |||
| Wild | 143 | 43 | 100 | |
| Mutation | 13 | 2 | 11 | |
| PIK3CA | 0.998 | |||
| Wild | 132 | 38 | 94 | |
| Mutation | 24 | 7 | 17 |
MAGE-C1, melanoma antigen gene C1; HER2, human epidermal growth factor receptor 2; KRAS, Kirsten rat sarcoma; BRAF, v-RAF murine sarcoma viral oncogene homolog B1; PIK3CA, Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha.
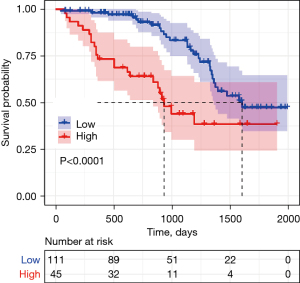
Kaplan-Meier survival analysis showed that the median overall survival duration of patients with high and low MAGE-C1 expression was 31.1 and 53.5 months, respectively, and the difference was statistically significant (P<0.001), as shown in Figure 3. MAGE-C1 outlier expression was positively associated with reduced survival duration.
Univariate and multivariate Cox regression analysis suggested that, compared to CRC patients with MAGE-C1 high expression, patients with MAGE-C1 low expression had better prognosis [hazard ratio (HR) =0.34, P<0.001]; compared to CRC patients with KRAS and BRAF mutations, patients with KRAS wild type (HR =0.31, P<0.001) and BRAF wild type (HR =0.32, P<0.05) had better prognosis. Compared to CRC patients with T3/T4 stage, patients with T1/T2 stage had a better prognosis (HR =0.22, P<0.05); compared to CRC patients within well or moderate differentiation, patients within poor differentiation had a worse prognosis (HR =2.21, P<0.05), as shown in Table 2.
Table 2
| Characteristics | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | ||
| Age (≤60/>60 years) | 1.57 | 0.86–2.88 | 0.144 | 1.11 | 0.56–2.17 | 0.771 | |
| Gender (male/female) | 0.68 | 0.39–1.18 | 0.172 | 0.84 | 0.44–1.60 | 0.590 | |
| T stage (T1 & T2/T3 & T4) | 0.11 | 0.04–0.32 | 0.001 | 0.22 | 0.07–0.67 | 0.007 | |
| N stage (N0/N1 & N2) | 0.24 | 0.13–0.42 | 0.001 | 0.69 | 0.34–1.41 | 0.307 | |
| M stage (M0/M1) | 0.19 | 0.11–0.34 | 0.001 | 0.52 | 0.25–1.05 | 0.069 | |
| Differentiation (poorly/well and moderately) | 3.60 | 2.02–6.43 | 0.001 | 2.21 | 1.04–4.68 | 0.038 | |
| MAGEC1 (low/high) | 0.34 | 0.20–0.60 | 0.001 | 0.34 | 0.18–0.64 | 0.001 | |
| HER2 (wild/mutation) | 0.51 | 0.16–1.63 | 0.255 | ||||
| KRAS (wild/mutation) | 0.46 | 0.26–0.80 | 0.006 | 0.31 | 0.16–0.61 | 0.001 | |
| BRAF (wild/mutation) | 0.46 | 0.22–0.94 | 0.032 | 0.32 | 0.14–0.75 | 0.008 | |
| PIK3CA (wild/mutation) | 0.62 | 0.31–1.23 | 0.173 | 0.80 | 0.38–1.69 | 0.562 | |
The characteristics with P≤0.2 in univariate analysis were included in the multivariate analysis. MAGE-C1, melanoma antigen gene C1; HER2, human epidermal growth factor receptor 2; KRAS, Kirsten rat sarcoma; BRAF, v-RAF murine sarcoma viral oncogene homolog B1; PIK3CA, Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha.
MAGE-C1 combined with clinicopathological characteristics and hotspot gene mutations in predicting survival rate of patients
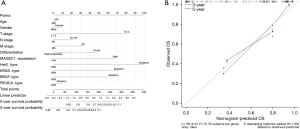
We designed a model that used the clinicopathological characteristics of patients and the information about CRC hotspot gene mutations to predict patient survival. First, the Cox regression model was employed to weight the scores of various factors and assign values to each factor, and then these values were added to acquire the total value. Next, we used this total value to calculate the patient’s corresponding predicted survival time in the model. The model is shown in Figure 4A. Finally, we tested the reliability and validity of the model. The results showed that the calibration chart was consistent with the primary cohort. The 3-year and 5-year OS nominal map predictions and actual observations were basically in line with those of the International Association for the Study of Lung Cancer (IASLC) verification cohort, as shown in Figure 4B.
Discussion
The worldwide incidence and mortality of CRC rank third and second among all cancers, respectively. There are about 1.8 million new cases reported and about 881 thousand people died due to CRC in 2018 (19). Survival rates for CRC patients can vary based on several factors. For some patients at an early stage, surgical removal of these tumors can sometimes eliminate cancer, which can improve the 5-year survival rate for these patients (20). In terms of late-stage patients, standard radiation therapy, chemotherapy, and targeted therapy have prolonged the survival time of patients and improved the quality of life of patients. Moreover, tumor immunotherapy has gained popularity, and its application, such as in the form of the checkpoint inhibitors of pembrolizumab and nivolumab, is being gradually broadened in clinical practice (21). Some studies have highlighted the role of immunotherapy in specific types of CRC (22). MAGE proteins, as targets for cancer immunotherapy, have also attracted increasing interest among researchers. Indeed, studies have shown that the MAGE protein could be used for immunotherapy and targeted therapy (23).
MAGE-C1, a CTA, is widely expressed in multiple myeloma. More than 85% of symptomatic patients with multiple myeloma show expression of MAGE-C1 in the bone marrow and peripheral blood (24). Furthermore, MAGE-C1 could be used to determine the types of malignant cells in multiple myeloma (13), and targeting MAGE-C1/CT7 expression has been shown to increase cell sensitivity to the proteasome inhibitor bortezomib (25). MAGE-C1 is expressed in a certain subset of ovarian carcinomas with no expression in borderline tumors or ovarian carcinomas of mucinous histology (15). Although the effect of some MAGE protein expression on patient survival has been studied, there is a lack of ample research on the effect of MAGE-C1 protein on the prognosis of CRC patients.
Our analysis of 156 cases with CRC showed that patients with high MAGE-C1 expression presented a worse prognosis than those with low expression. Overexpression of MAGE family proteins has been associated with the poor prognosis of many cancers in several studies. For example, MAGE-A12 was related to the high TNM staging of gastric cancer, and was demonstrated to be an independent indicator of poor prognosis in patients with gastric cancer (26). The expression of MAGE-A1-6 in the peritoneal lavage after gastric cancer surgery is correlated with the patient’s reduced disease-free survival rate, indicating its association with tumor recurrence (27). MAGE family proteins were also found to be related to other solid tumors. For example, MAGE-A3 was shown to be a poor prognostic indicator of non-small cell lung cancer (28), MAGE-A3 and A6 were found to be correlated to the poor prognosis of specific types of breast cancer (7), while MAGE-A1 and A10 were associated with the poor prognosis of ovarian cancer (29). However, the impact of MAGE-C1 protein on the prognosis of CRC has not been extensively reported.
This study observed that the high expression of MAGE-C1 in CRC was related to higher TNM staging, which is similar to the relationship between MAGE-A12 and TNM staging in gastric cancer (26). The increase in TNM staging indicated tumor progression to a higher degree and also suggested MAGE-C1 functions to be a driver of tumorigenesis in CRC. Studies have demonstrated that cell infiltration with high expression of MAGE-C2 to be increased in cell lines cultured in vitro (30).
The bias produced by a single indicator to predict the survival of cancer patients is often large (31). Multiple indicators to predict the survival of CRC often produce better results. According to the expression level of MAGE-C1, patient’s clinicopathological characteristics, genes with clinical concern, and patient survival data, we designed a model that integrated the clinicopathological characteristics and genes with clinical concern of patients with CRC to predict their survival rate. Although the model still needs further improvement, it can be used as an important tool for the clinicopathological evaluation of patient prognosis. Compared with other methods, our model may be more amenable to clinicians because this method only needs to collect clinicopathological characteristics and genes with clinical concern of patients, which are genes recommended for testing in clinical CRC diagnosis and treatment guidelines. The calculation method of the patient’s expected survival is simple and can be integrated into one program. Thus overall, the method is relatively time-saving and efficient.
This study has some limitations. The data collected were only from 156 patients with CRC. Only 7 clinicopathological parameters and 6 genes with clinical concern were introduced into the prediction model. Thus, the accuracy and effectiveness of the model need to be further validated. Besides, we are not clear how MAGE-C1 affects CRC progression, and that’s also what we need to do next.
Conclusions
Overall, MAGE-C1 is a poor prognostic indicator of CRC. Its pathogenicity in CRC needs to be further studied. Compared with using a single index to predict the prognosis of CRC, it is more effective in predicting the prognosis of CRC through its use of a mathematical model that incorporates multiple parameters.
Acknowledgments
The authors thank Yuan Yin of the Wuxi Cancer Institute, Affiliated Hospital of Jiangnan University for her constructive suggestions.
Funding: This work was funded by the Science and Technology Plan of Wuxi Municipal Health Commission (No. T202114) and Natural Science Foundation of Fujian Province (No. 2021J05276).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-739
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-739
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-739). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Affiliated Hospital of Jiangnan University (No. 2014-012-001). All patients gave written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Simpson AJ, Caballero OL, Jungbluth A, et al. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005;5:615-25. [Crossref] [PubMed]
- Ruiz R, Hunis B, Raez LE. Immunotherapeutic agents in non-small-cell lung cancer finally coming to the front lines. Curr Oncol Rep 2014;16:400. [Crossref] [PubMed]
- Bai S, He B, Wilson EM. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol Cell Biol 2005;25:1238-57. [Crossref] [PubMed]
- Bai S, Wilson EM. Epidermal-growth-factor-dependent phosphorylation and ubiquitinylation of MAGE-11 regulates its interaction with the androgen receptor. Mol Cell Biol 2008;28:1947-63. [Crossref] [PubMed]
- Bhatia N, Xiao TZ, Rosenthal KA, et al. MAGE-C2 promotes growth and tumorigenicity of melanoma cells, phosphorylation of KAP1, and DNA damage repair. J Invest Dermatol 2013;133:759-67. [Crossref] [PubMed]
- Kim YD, Park HR, Song MH, et al. Pattern of cancer/testis antigen expression in lung cancer patients. Int J Mol Med 2012;29:656-62. [Crossref] [PubMed]
- Ayyoub M, Scarlata CM, Hamaï A, et al. Expression of MAGE-A3/6 in primary breast cancer is associated with hormone receptor negative status, high histologic grade, and poor survival. J Immunother 2014;37:73-6. [Crossref] [PubMed]
- Karpf AR, Bai S, James SR, et al. Increased expression of androgen receptor coregulator MAGE-11 in prostate cancer by DNA hypomethylation and cyclic AMP. Mol Cancer Res 2009;7:523-35. [Crossref] [PubMed]
- Mori M, Inoue H, Mimori K, et al. Expression of MAGE genes in human colorectal carcinoma. Ann Surg 1996;224:183-8. [Crossref] [PubMed]
- Shantha Kumara HM, Grieco MJ, Caballero OL, et al. MAGE-A3 is highly expressed in a subset of colorectal cancer patients. Cancer Immun 2012;12:16. [PubMed]
- Zhang QM, He SJ, Shen N, et al. Overexpression of MAGE-D4 in colorectal cancer is a potentially prognostic biomarker and immunotherapy target. Int J Clin Exp Pathol 2014;7:3918-27. [PubMed]
- Zhan W, Zhang Z, Zhang Y, et al. Prognostic value of MAGE-A9 expression in patients with colorectal cancer. Clin Res Hepatol Gastroenterol 2016;40:239-45. [Crossref] [PubMed]
- Wienand K, Shires K. The use of MAGE C1 and flow cytometry to determine the malignant cell type in multiple myeloma. PLoS One 2015;10:e0120734. [Crossref] [PubMed]
- Xu P, Zhang L, Wang X, et al. Expression of MAGE-C1/CT7 provides prognostic information in multiple myeloma. Leuk Lymphoma 2017;58:244-6. [Crossref] [PubMed]
- Zimmermann AK, Imig J, Klar A, et al. Expression of MAGE-C1/CT7 and selected cancer/testis antigens in ovarian borderline tumours and primary and recurrent ovarian carcinomas. Virchows Arch 2013;462:565-74. [Crossref] [PubMed]
- Hou S, Sang M, Zhao L, et al. The expression of MAGE-C1 and MAGE-C2 in breast cancer and their clinical significance. Am J Surg 2016;211:142-51. [Crossref] [PubMed]
- Li R, Qu H, Wang S, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018;34:2515-7. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chan GHJ, Chee CE. Making sense of adjuvant chemotherapy in colorectal cancer. J Gastrointest Oncol 2019;10:1183-92. [Crossref] [PubMed]
- Wang C, Sandhu J, Fakih M. Complete response to pembrolizumab in a patient with metastatic colon cancer with microsatellite instability and a history of Guillain-Barre syndrome. J Gastrointest Oncol 2019;10:161-5. [Crossref] [PubMed]
- Kalyan A, Kircher S, Shah H, et al. Updates on immunotherapy for colorectal cancer. J Gastrointest Oncol 2018;9:160-9. [Crossref] [PubMed]
- Weon JL, Potts PR. The MAGE protein family and cancer. Curr Opin Cell Biol 2015;37:1-8. [Crossref] [PubMed]
- Tinguely M, Jenni B, Knights A, et al. MAGE-C1/CT-7 expression in plasma cell myeloma: sub-cellular localization impacts on clinical outcome. Cancer Sci 2008;99:720-5. [Crossref] [PubMed]
- de Carvalho F, Costa ET, Camargo AA, et al. Targeting MAGE-C1/CT7 expression increases cell sensitivity to the proteasome inhibitor bortezomib in multiple myeloma cell lines. PLoS One 2011;6:e27707. [Crossref] [PubMed]
- Wu J, Wang J, Shen W. Identification of MAGEA12 as a prognostic outlier gene in gastric cancers. Neoplasma 2017;64:238-43. [Crossref] [PubMed]
- Jeon CH, Kim IH, Chae HD. Prognostic value of genetic detection using CEA and MAGE in peritoneal washes with gastric carcinoma after curative resection: result of a 3-year follow-up. Medicine (Baltimore) 2014;93:e83. [Crossref] [PubMed]
- Gure AO, Chua R, Williamson B, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res 2005;11:8055-62. [Crossref] [PubMed]
- Daudi S, Eng KH, Mhawech-Fauceglia P, et al. Expression and immune responses to MAGE antigens predict survival in epithelial ovarian cancer. PLoS One 2014;9:e104099. [Crossref] [PubMed]
- Yang F, Zhou X, Miao X, et al. MAGEC2, an epithelial-mesenchymal transition inducer, is associated with breast cancer metastasis. Breast Cancer Res Treat 2014;145:23-32. [Crossref] [PubMed]
- Mahar AL, Compton C, Halabi S, et al. Personalizing prognosis in colorectal cancer: A systematic review of the quality and nature of clinical prognostic tools for survival outcomes. J Surg Oncol 2017;116:969-82. [Crossref] [PubMed]
(English Language Editor: J. Gray)


