
The effect of PPP2CA expression on the prognosis of patients with hepatocellular carcinoma and its molecular biological characteristics
Introduction
Hepatocellular carcinoma (HCC) is a malignant tumor with the highest cancer-related mortality in the world (1,2). The occurrence and invasion of HCC involve the activity and interaction of multiple genes. Protein phosphatase 2A (PP2A) is a serine/threonine phosphatase highly expressed in eukaryotes. It is composed of structural subunit A (PR65), catalytic subunit C (PP2Ac), and a complex composed of various variable regulatory subunits B. The PPP2CA gene encodes the α subtype of the PP2A catalytic subunit and regulates PP2A activity by selecting the PP2A regulatory subunit, thereby playing a vital role in the activity of PP2A (3-5). Depending on the setting, PP2A can act as a tumor suppressor or promoter (6,7), but whether PP2A plays an inhibitory or promoting role in HCC has remained controversial. A large number of studies have proved that PP2A is a tumor suppressor in HCC, which mainly includes the following three aspects: PP2A endogenous inhibitor; targeted inhibition of PP2A; PP2A expression of different subunits decrease (8). Kong et al. (9) found that jigging acid promotes the occurrence and invasion of HCC by inhibiting the activity of PP2A and promoting the process of epithelial-mesenchymal transition. This shows that PP2A plays an inhibitory role in HCC; however, Gong et al. (10) was found that the up-regulation of PP2Ac contributes to the aggressiveness of HCC. Therefore, whether PP2A is a tumor suppressor or a promoter in HCC is still controversial.
In this study, we will combine clinical, immunohistochemistry, and bioinformatics to explore the role of PPP2CA in HCC. We retrospectively analyzed 128 HCC patients who underwent surgery for the first time in the Department of Hepatobiliary Surgery, Cancer Hospital of Guangxi Medical University from July 2017 to July 2019, and compared the expression of PPP2CA in HCC tissues and non-HCC tissues. We also compared the clinical data and survival of patients with low or high PPP2CA expression using bioinformatics and public databases, the PPP2CA expression between HCC tissues and non-HCC tissues, and verified the survival rate of patients with low or high PPP2CA expression. Finally, we employed various bioinformatics programs to explore the possible pathways and targets of PPP2CA’s participation in HCC.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-720).
Methods
Data collection
Immunohistochemical (IHC) tests were performed on liver cancer tissues and matched non-liver cancer tissue samples of 128 HCC patients who were treated at the Department of Hepatobiliary Surgery, Cancer Hospital of Guangxi Medical University from July 2017 to July 2019. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by board of Ethics Committee of Guangxi Medical University Cancer Hospital (No. LW2021097) and informed consent was taken from all the patients. Tissue samples were included for analysis based on the following participant inclusion criteria: (I) underwent radical hepatocarcinoma resection; (II) clearly diagnosed as HCC based on postoperative pathology; (III) did not receive any anti-cancer treatment before surgery; (IV) did not have any malignant tumor other than HCC.
Clinical data were collected including the patient’s gender, age, family history of liver cancer, Child-Pugh grade, presence or absence of liver cirrhosis, alpha-fetoprotein (AFP), tumor size, tumor number, Barcelona clinic liver cancer (BCLC) stage, total bilirubin (TBil), aspartate aminotransferase (AST), alanine aminotransferase (ALT), albumin (ALB), prothrombin time (PT), surgical resection method, degree of tumor differentiation, whether the capsule was intact, whether there was microvascular invasion (MVI), liver capsule invasion, lymph node metastasis, portal cancer thrombus, bile duct cancer thrombus, P-53, Ki-67, and so on.
Follow-up
All participants were followed up as desired to their time of death, loss to follow-up, or until July 2021. Survival data was drawn from patient medical records and telephone follow-up. Recurrence-free survival (RFS) was defined as the time from surgery to recurrence or death. Overall survival (OS) was defined as the time from surgery to death. Both RFS and OS were defined as the time from the day of surgery to the discovery of tumor recurrence or the last follow-up, that is, July 2021.
IHC
The HCC specimens and matched non-HCC tissues were fixed with 10% formalin solution, embedded in paraffin, and cut into slices about 4 µm thick. Sections were deparaffinized in xylene at 37 °C, washed in a gradient ethanol series, incubated in 10 mmol/L citrate buffer (pH 6.0) for antigen retrieval, rinsed in phosphate-buffered saline (PBS), incubated in 0.3% H2O2 for 10 min at room temperature to inhibit endogenous peroxidase, and then rinsed again with PBS. The sections were incubated with anti-PPP2CA antibody (1:1,000) (ab106262; Abcam, Cambridge, UK) at 37 °C for 1 h, and then rinsed thoroughly in PBS. The sections were incubated with MaxVision™ (Shenzhen, China)/horseradish peroxidase for 30 min at room temperature, washed with PBS, and stained with diamino diphenyl and hematoxylin. The slices were dehydrated by a series of graded ethanol, dried, and sealed with neutral gum. At the same time, retinal tissue was treated as a positive control, while HCC tissue was treated with PBS instead of the primary antibody as a negative control.
The IHC staining used 2-component scoring for semi-quantification. First, the intensity of staining was assigned 0 points (uncolored), 1 point (light yellow), 2 points (yellow), or 3 points (dark yellow). Second, the percentage of the total number of observed cells that are stained was assigned as 1 point (≤25%), 2 points (26–50%), 3 points (51–75%), or 4 points (>75%). The 2 scores were multiplied to attain a total score of 0–12. A total score of 0–4 was considered “low expression”, and a score of 5–12 was considered “high expression”. All slides were evaluated by 2 senior pathologists in double-blind evaluation. If the results of the 2 pathologists were inconsistent, the review was repeated.
The clinical significance of PPP2CA in HCC
We stratified HCC patients according to PPP2CA expression and analyzed the relationship between PPP2CA expression and clinical outcome. We used the Kaplan-Meier plotter to compare the OS and RFS of patients with high or low PPP2CA expression.
Exploration of PPP2CA’s potential routes of participation in HCC
Using the GSE76427 data set, we explored genes that may be related to PPP2CA and examined candidate differentially expressed genes (DEGs) in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) databases in terms of biological processes, molecular functions, and cellular components. The GO and KEGG analyses were performed using the DAVID tool (https://david.ncifcrf.gov/) (11). The results of GO and KEGG analysis were depicted in bubble charts. The online Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) was used to construct a protein-protein interaction (PPI) network. The PPI network of DEGs was constructed from the STRING database and visualized by Cytoscape software (https://cytoscape.org/).
Statistical analysis
The statistical software SPSS 26.0 (IBM Corp., Armonk, NY, USA) was used to analyze participant data: count data was compared with the chi-square test; the Shapiro-Wilk test was used to determine the normality of measurement data, and independent sample t-test was used for data conforming to a normal distribution and homogeneous variance. The mean ± standard deviation () was used for comparison; non-normally distributed data were compared using the nonparametric rank-sum test Mann-Whitney U test, and the results were expressed in median and quartile. A P value <0.05 indicated a statistically significant difference. The Kaplan-Meier method was used for survival analysis and comparison.
Results
The expression of PPP2CA in liver cancer tissues
The experimental results of IHC showed that PPP2CA was highly expressed in most HCC tissues, but low in most non-HCC tissues, and was mainly expressed in the cytoplasm (Figure 1).
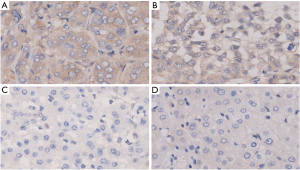
The clinical application of PPP2CA in HCC
The general clinical data of the 2 groups of participants were compared (Table 1). Among them, the differences in Child-Pugh classification, AST, MVI, and portal cancer thrombus were statistically significant (all P<0.05). Gender, age, family history of liver cancer, Child-Pugh grade, liver cirrhosis, AFP, tumor size, tumor number, BCLC staging, TBil, ALT, ALB, PT, surgical resection method, tumor differentiation degree, whether the capsule was intact, whether there were no statistically significant differences in envelope invasion, lymph node metastasis, bile duct tumor thrombus, P-53, Ki-67, and so on were all also statistically significant (all P>0.05).
Table 1
| Parameters | PPP2CA expression | P value | χ2/Z | |
|---|---|---|---|---|
| High expression (n=88) | Low expression (n=40) | |||
| Gender | 0.114 | 2.495 | ||
| Male | 76 | 30 | ||
| Female | 12 | 10 | ||
| Age, years | 0.694 | 0.155 | ||
| ≤50 | 56 | 24 | ||
| >50 | 32 | 16 | ||
| Family history of HCC | 0.255 | 1.298 | ||
| No | 67 | 34 | ||
| Yes | 21 | 6 | ||
| Child-Pugh classification | 0.041* | 4.173 | ||
| A | 84 | 34 | ||
| B | 4 | 6 | ||
| Liver cirrhosis | 0.237 | 1.396 | ||
| No | 30 | 18 | ||
| Yes | 58 | 22 | ||
| AFP, ng/mL | 0.535 | 0.384 | ||
| >400 | 41 | 21 | ||
| ≤400 | 47 | 19 | ||
| Tumor size (cm) | 0.623 | 0.242 | ||
| ≤5 | 28 | 11 | ||
| >5 | 60 | 29 | ||
| Number of tumors | 0.537 | 0.381 | ||
| Single | 62 | 26 | ||
| Multiple | 26 | 14 | ||
| BCLC-staging | 0.564 | 1.144 | ||
| A | 39 | 14 | ||
| B | 13 | 8 | ||
| C | 36 | 18 | ||
| TBil | 15 (15.8–30.7) | 16.4 (15.7–21.3) | 0.309 | 0.965 |
| ALT | 39.5 [44–65] | 34.5 [33–60] | 0.471 | 0.846 |
| AST | 47 [50–64] | 37 [41–67] | 0.018* | 1.537 |
| ALB | 36.4 (35.6–37.2) | 36.2 (35.7–37.9) | 0.870 | 0.596 |
| PT | 12.2 (12.0–12.6) | 12.3 (12.0–12.8) | 0.977 | 0.477 |
| Surgical resection method | 0.787 | 0.073 | ||
| ≤2 liver segment | 64 | 30 | ||
| >3 liver segment | 24 | 10 | ||
| Tumor differentiation | 0.848 | 0.037 | ||
| Poor differentiation | 38 | 18 | ||
| Well differentiated | 50 | 22 | ||
| Envelope intact | 0.365 | 0.821 | ||
| Yes | 43 | 17 | ||
| No | 45 | 23 | ||
| MVI | 0.001* | 10.670 | ||
| No | 28 | 25 | ||
| Yes | 60 | 15 | ||
| Liver capsule invasion | 1.000 | 0.000 | ||
| No | 33 | 15 | ||
| Yes | 55 | 25 | ||
| Lymph node metastasis | 0.293 | 3.724 | ||
| No | 77 | 33 | ||
| Yes | 9 | 6 | ||
| P-53 | 0.715 | 0.133 | ||
| Negative | 35 | 15 | ||
| Positive | 53 | 25 | ||
| Ki-67 | 0.737 | 0.112 | ||
| ≤50 | 49 | 21 | ||
| >50 | 39 | 19 | ||
| Portal vein tumor thrombus | 0.018* | 5.623 | ||
| No | 63 | 20 | ||
| Yes | 25 | 20 | ||
| Bile duct tumor thrombus | 0.857 | 0.025 | ||
| No | 83 | 38 | ||
| Yes | 5 | 2 | ||
*, P<0.05. HCC, hepatocellular carcinoma; AFP, alpha fetoprotein; BCLC, Barcelona clinic liver cancer; Tbil, total bilirubin; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALB, albumin; PT, prothrombin time.
In addition, the Kaplan-Meier survival curve showed that the low expression group had better OS (P=0.04) and RFS (P=0.019) (Figure 2).
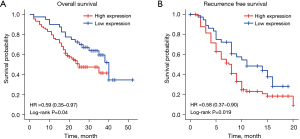
The expression of PPP2CA in HCC and non-HCC tissues in The Cancer Genome Atlas database
We used the GSE76427 data set to compare expression of the PPP2CA gene in HCC tissues and non-HCC tissues. The results showed that the expression level of PPP2CA in HCC tissues was higher than that in non-HCC tissues (Figure 3), and the difference was statistically significant (P=0.006).
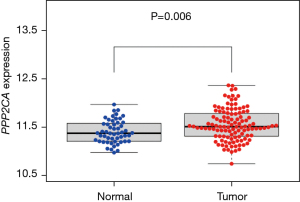
The correlation between PPP2CA gene and HCC
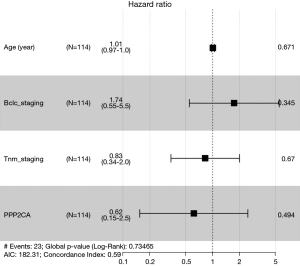
We divided the groups by the median and third quartile of PPP2CA gene levels and compared the OS of the 2 participant groups, which revealed that there was no significant difference in the OS of the 2 groups (P>0.05) (Figure 4). Correlation analysis of clinical parameters showed that PPP2CA has little correlation with HCC patients’ age and BCLC-stage (P>0.05), but it is significantly correlated with tumor-node-metastasis (TNM)-stage and gender (P<0.05) (Figure 5). Univariate Cox regression and multivariate COX regression analysis showed that age, BCLC-stage, TNM-stage, and PPP2CA are not factors that affect the survival of HCC patients (P>0.05) (Table 2, Figure 6).
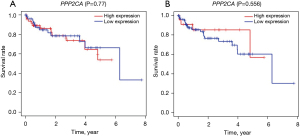
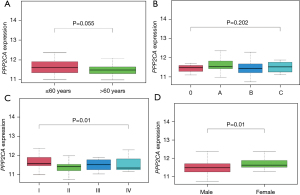
Table 2
| Univariate COX regression | Multivariate cox regression | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | HR.95CL | HR.95H | P value | HR | HR.95CL | HR.95H | P value | ||
| Age (years) | 1.011052 | 0.976926 | 1.04637 | 0.530383 | 1.007871 | 0.972107 | 1.044951 | 0.670609 | |
| BCLC-staging | 1.393437 | 0.732097 | 2.6522 | 0.312338 | 1.742795 | 0.550207 | 5.520351 | 0.345013 | |
| TNM-staging | 1.163814 | 0.700212 | 1.93436 | 0.558405 | 0.82606 | 0.34265 | 1.991467 | 0.670387 | |
| PPP2CA | 0.607936 | 0.16353 | 2.260051 | 0.457558 | 0.6164 | 0.153898 | 2.468829 | 0.494327 | |
OS, overall survival; HCC, hepatocellular carcinoma; BCLC, Barcelona clinic liver cancer; TNM, tumor-node-metastasis; HR, hazard ratio.
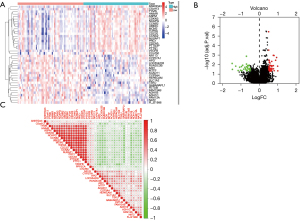
PPP2CA and its DEGs
We analyzed HCC sample tissues and non-HCC control sample tissues from the GSE76427 data set. The PPP2CA gene and 71 DEGs were identified between HCC sample tissues and non-HCC control sample tissues, and they were drawn by heat map package and R software. The heat map (Figure 7A) and volcano map (Figure 7B) of the first 40 DEGs (the first 20 up-regulated DEGs and the first 20 down-regulated DEGs) were also analyzed, and the correlation between PPP2CA and DEGs was analyzed (Figure 7C).
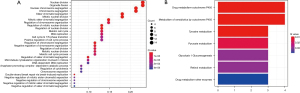
GO analyses and KEGG pathways analyses
We performed enrichment analysis of the GO and KEGG pathways of PPP2CA and its DEGs. The GO analysis results showed that the main functions of biological processes are nuclear division, organelle fission, nuclear chromosome separation, and chromatid separation (Figure 8A). The most important of the 7 KEGG pathways identified was drug metabolism-cytochrome P450, followed by the metabolism of cytochrome P450 to xenobiotics (Figure 8B).
PPI network of PPP2CA and its DEGs and identification of key gene
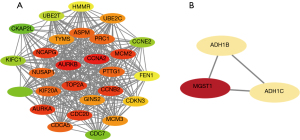
We uploaded PPP2CA and its DEGs to STRING to construct a PPI network (Figure 9A). The PPI network was optimized using Cytoscape software to obtain a clearer PPI network (Figure 9B) and 2 sub-networks (Figure 10). The results showed that CCNA2, AURKB, TOP2A, NCAPG, MCM2, CDC20, CCMB2, AURKA, and MGST1 were the top 9 highly connected hub genes.

Discussion
The PPP2CA gene encodes the α subtype of the PP2A catalytic subunit, which regulates PP2A activity by selecting PP2A regulatory subunits (12). In previous studies, the up-regulation of PPP2CA was considered to be related to the poor prognosis of triple-negative breast cancer (13) and pancreatic cancer (14). High expression of PPP2CA is also related to glioma, and mir-130b-ceRNA can promote the epithelial-mesenchymal transition and invasion of glioma by targeting PPP2CA (15). In addition, the results of a phase I study showed that the anti-tumor drug LB-100 combined with existing anti-tumor drugs has better efficacy in the treatment of pancreatic cancer, glioma, and other diseases than single-agent efficacy, and supported that LB-100 is specific inhibit PPP2CA to improve efficacy (16). On the contrary, colorectal patients with low expression of PPP2CA have poor OS, and low expression of PPP2CA is significantly related to the later TNM stage (17). Some studies have shown that decreased expression of PPP2CA can increase the susceptibility of Chinese people to gastric cancer (18). The above studies showed that PPP2CA plays different roles in different cancer settings, and there may be multiple mechanisms and pathways that cause this phenomenon. The role of PP2A in HCC is still controversial (9,19,20) and research of PPP2CA in the treatment of HCC is valuable.
We used IHC to detect the expression of PPP2CA in HCC tissues and non-HCC tissues. We found that PPP2CA is highly expressed in most HCC tissues, and its expression is mainly distributed in the cytoplasm. Duong et al. (21) found that the expression of PP2Ac in HCC tissues was significantly higher than that in non-HCC tissues from the same patient. We divided the expression of PPP2CA into a high expression group (medium positive and + strong positive) and a low expression group (negative and weak positive expression) according to the results of IHC detection. By comparing the clinical data of the 2 groups, we found that the high expression of PPP2CA and MVI is significantly related to portal vein tumor thrombus. In addition, participants in the PPP2CA high expression group had worse OS and RFS. These clinical data indicate that PPP2CA may promote the invasion and recurrence of HCC to a certain extent. In a WARD study, it was also found that PP2A promoted the invasion of HCC through extracellular remodeling (22), which is consistent with our speculation. In addition, some studies have shown that the high expression of PPP2CA has been associated with hepatitis B (HBV) or hepatitis C (HCV) viral infection (23). However, since most of this study is HBV-related HCC, verification would require much larger data and multi-center studies.
In addition, we used the GSE76427 dataset to analyze expression of the PPP2CA gene in HCC tissues and non-HCC tissues and found that the expression level of PPP2CA in HCC tissues was significantly higher than that in non-liver cancer tissues. Kaplan-Meier survival analysis showed that the expression of PPP2CA is not different from the OS of HCC patients, but the high expression of PPP2CA is significantly related to the TNM stage of HCC patients. Combined with the results of clinical studies, we can infer that PPP2CA plays an important role in the invasion and metastasis of HCC. However, in the end, multivariate regression analysis found that the PPP2CA gene may not be an independent risk factor for poor prognosis in HCC patients. In our clinical studies, participants in the PPP2CA high expression group had worse OS and RFS. At the same time, Gong et al. (10) showed that the up-regulation of PP2Ac expression has a negative impact on the OS of HCC patients, and the overexpression of PP2Ac is an independent risk factor for poor prognosis of HCC patients. Therefore, we have reason to believe that PP2A plays a role in promoting HCC. In addition, we found that the expression of PPP2CA is significantly related to gender in the analysis of bioinformatics research, but in clinical research, we did not find that PPP2CA is related to gender. This may be related to the fact that the number of cases we collected in the clinic was small, and more cases need to be collected and further analyzed. In addition, PPP2CA is highly expressed in HCC tissues. Our clinical studies have also shown that the expression of PPP2CA is significantly related to the RFS and OS of HCC patients. However, through the analysis of the GSE76427 data set, we found that the prognosis of HCC patients with different PPP2CA expressions is not different. Yes, this may be related to the insufficient number of cases in our data set, but in our previous research (24), we analyzed data from the cancer genome map database, gene expression comprehensive database, and our internal RNA-Seq database. The control data of 2,545 HCC patients and 1,993 non-HCC patients found that the expression of PPP2CA in HCC tissues was significantly higher than that in non-HCC tissues, and immunohistochemistry of tissue sections also confirmed that PPP2CA protein was up-regulated in HCC tissues; in addition, HCC patients shorter OS and DFS are all related to the high expression of PPP2CA.
We performed KEGG enrichment analysis on PPP2CA and its DEGs, revealing that the most important of the 7 KEGG pathways identified were drug metabolism-cytochrome P450, followed by the metabolism of cytochrome P450 to xenobiotics. These metabolic processes and the pathway provide evidence that pivot genes may be involved in the occurrence of HCC. The occurrence of HCC involves the up-regulation of drug metabolism-cytochrome P450 pathway and related genes (25), and is also related to poor OS. Therefore, we speculate that PPP2CA may affect HCC through the metabolism of cytochrome P450 and cytochrome P450 on xenobiotics. In addition, our PPI network further identified CCNA2 as an up-regulated central gene of HCC, which is consistent with the study of Song et al. (26), that is, increased messenger RNA (mRNA) expression of the CCNA2 gene is related to unfavorable RFS and OS in HCC patients. In addition, studies have also shown that AURKB has an important predictive value for the prognosis of HCC (27,28).
Our research also found that high expression of the TOP2A, NCAPG, MCM2, CDC20, CCMB2, AURKA, and MGST1 genes has a negative impact on the prognosis of HCC. This requires further experimental research and the identification of targeted drugs for these hub genes to determine their clinical applications, explore their biological behaviors, such as invasion, metastasis, and proliferation capabilities, and verify the impact of potential drugs on their targeted genes.
In conclusion, our research showed that PPP2CA is highly expressed in liver cancer tissues and is significantly related to the gender and TNM staging of liver cancer patients. Through the analysis of differential genes, high PPP2CA expression is associated with poor prognosis of HCC patients, GO and KEGG pathway analysis, it was found that PPP2CA may act on liver cancer through multiple targets and multiple pathways, PPP2CA plays a role in promoting HCC. The study of PPP2CA may be a potential therapeutic target for liver cancer, and the study of the effect and mechanism of PPP2CA on liver cancer has great potential value for the research of new drugs for liver cancer.
Acknowledgments
Funding: This study was supported by the Suitable Medical and Health Technology Research in Developmental Projects of Guangxi (No. S201516), and a Project of the Guangxi Province Education Commission of the Teacher Foundation for Youth (No. KY2016YB085).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-720
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-720
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-720). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013) and were approved by the Ethics Committee of Guangxi Medical University Cancer Hospital (No. LW2021097), and all participants provided their written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Petrick JL, Florio AA, Znaor A, et al. International trends in hepatocellular carcinoma incidence, 1978-2012. Int J Cancer 2020;147:317-30. [Crossref] [PubMed]
- Fernandes ES, Rodrigues PD, Álvares-da-Silva MR, Scaffaro LA, Farenzena M, Teixeira UF, Waechter FL. Treatment strategies for locally advanced hepatocellular carcinoma. Transl Gastroenterol Hepatol 2019;4:12. [Crossref] [PubMed]
- Yoon S, Kook T, Min HK, et al. PP2A negatively regulates the hypertrophic response by dephosphorylating HDAC2 S394 in the heart. Exp Mol Med 2018;50:1-14. [Crossref] [PubMed]
- Kauko O, Westermarck J. Non-genomic mechanisms of protein phosphatase 2A (PP2A) regulation in cancer. Int J Biochem Cell Biol 2018;96:157-64. [Crossref] [PubMed]
- Raman D, Pervaiz S. Redox inhibition of protein phosphatase PP2A: Potential implications in oncogenesis and its progression. Redox Biol 2019;27:101105. [Crossref] [PubMed]
- Dzulko M, Pons M, Henke A, et al. The PP2A subunit PR130 is a key regulator of cell development and oncogenic transformation. Biochim Biophys Acta Rev Cancer 2020;1874:188453. [Crossref] [PubMed]
- Shah VM, English IA, Sears RC. Select Stabilization of a Tumor-Suppressive PP2A Heterotrimer. Trends Pharmacol Sci 2020;41:595-7. [Crossref] [PubMed]
- Yang CL, Huang S, Zhang ZM. Mechanism of action of protein phosphatase 2A in the promotion and inhibition of hepatocellular carcinoma. J Clin Hepatol 2019;35:1123-8.
- Kong J, Li D, Zhang S, et al. Okadaic acid promotes epithelial-mesenchymal transition of hepatocellular carcinoma cells by inhibiting protein phosphatase 2A. J Cell Biochem 2020; Epub ahead of print. [Crossref] [PubMed]
- Gong SJ, Feng XJ, Song WH, et al. Upregulation of PP2Ac predicts poor prognosis and contributes to aggressiveness in hepatocellular carcinoma. Cancer Biol Ther 2016;17:151-62. [Crossref] [PubMed]
- Delker DA, Geter DR, Roop BC, et al. Oncogene expression profiles in K6/ODC mouse skin and papillomas following a chronic exposure to monomethylarsonous acid. J Biochem Mol Toxicol 2009;23:406-18. [Crossref] [PubMed]
- Longin S, Zwaenepoel K, Louis JV, et al. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J Biol Chem 2007;282:26971-80. [Crossref] [PubMed]
- Chen J, Liu C, Cen J, et al. KEGG-expressed genes and pathways in triple negative breast cancer: Protocol for a systematic review and data mining. Medicine (Baltimore) 2020;99:e19986. [Crossref] [PubMed]
- Hang J, Lau SY, Yin R, et al. The role of phosphoprotein phosphatases catalytic subunit genes in pancreatic cancer. Biosci Rep 2021;41:BSR20203282. [Crossref] [PubMed]
- Tong L, Chu M, Yan B, et al. MTDH promotes glioma invasion through regulating miR-130b-ceRNAs. Oncotarget 2017;8:17738-49. [Crossref] [PubMed]
- D'Arcy BM, Swingle MR, Papke CM, et al. The Antitumor Drug LB-100 Is a Catalytic Inhibitor of Protein Phosphatase 2A (PPP2CA) and 5 (PPP5C) Coordinating with the Active-Site Catalytic Metals in PPP5C. Mol Cancer Ther 2019;18:556-66. [Crossref] [PubMed]
- Yong L. YuFeng Z, Guang B. Association between PPP2CA expression and colorectal cancer prognosis tumor marker prognostic study. Int J Surg 2018;59:80-9. [Crossref] [PubMed]
- Huang T, He K, Mao Y, et al. Genetic variants in PPP2CA are associated with gastric cancer risk in a Chinese population. Sci Rep 2017;7:11499. [Crossref] [PubMed]
- Yu HC, Hung MH, Chen YL, et al. Erlotinib derivative inhibits hepatocellular carcinoma by targeting CIP2A to reactivate protein phosphatase 2A. Cell Death Dis 2014;5:e1359. [Crossref] [PubMed]
- Ciou SC, Chou YT, Liu YL, et al. Ribose-5-phosphate isomerase A regulates hepatocarcinogenesis via PP2A and ERK signaling. Int J Cancer 2015;137:104-15. [Crossref] [PubMed]
- Duong FH, Dill MT, Matter MS, et al. Protein phosphatase 2A promotes hepatocellular carcinogenesis in the diethylnitrosamine mouse model through inhibition of p53. Carcinogenesis 2014;35:114-22. [Crossref] [PubMed]
- Ward MP, Spiers JP. Protein phosphatase 2A regulation of markers of extracellular matrix remodelling in hepatocellular carcinoma cells: functional consequences for tumour invasion. Br J Pharmacol 2017;174:1116-30. [Crossref] [PubMed]
- Christen V, Duong F, Bernsmeier C, et al. Inhibition of alpha interferon signaling by hepatitis B virus. J Virol 2007;81:159-65. [Crossref] [PubMed]
- Yang CL, Qiu X, Lin JY, et al. Potential Role and Clinical Value of PPP2CA in Hepatocellular Carcinoma. J Clin Transl Hepatol 2021;9:661-71. [Crossref] [PubMed]
- Zhou L, Du Y, Kong L, et al. Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. Onco Targets Ther 2018;11:1861-9. [Crossref] [PubMed]
- Song X, Du R, Gui H, et al. Identification of potential hub genes related to the progression and prognosis of hepatocellular carcinoma through integrated bioinformatics analysis. Oncol Rep 2020;43:133-46. [PubMed]
- Moteleub HNAE, Moety AAAE, Baddour NM, et al. Predictors for hepatocellular carcinoma recurrence after microwave ablation. Clin Exp Hepatol 2020;6:77-84. [Crossref] [PubMed]
- Zhang Y, Yang L, Shi J, et al. The Oncogenic Role of CENPA in Hepatocellular Carcinoma Development: Evidence from Bioinformatic Analysis. Biomed Res Int 2020;2020:3040839. [Crossref] [PubMed]
(English Language Editor: J. Jones)


