
Long noncoding RNA NEAT1 changes exosome secretion and microRNA expression carried by exosomes in hepatocellular carcinoma cells
Introduction
Hepatocellular carcinoma (HCC) has the third-highest cancer-related mortality rate (1), as well as a high risk of recurrence and poor prognosis. However, there are currently few biomarkers for the diagnosis of early HCC. Despite their limited benefits, resection and liver transplant are the primary options for the treatment of HCC (2). HCC progression is not only determined by the biological behavior of tumor cells but is also influenced by the tumor microenvironment (TME) (3,4). Increasing evidence has suggested that the TME plays a vital role in HCC progression (3,5).
Exosomes are double-layered vesicles (approximately 30–150 nm in diameter) that contain nucleic proteins, acids, and lipids, which regulate intercellular communication (6). Numerous studies have suggested that exosomes contribute to HCC progression by affecting chemoresistance, angiogenesis, metastasis, and the immune response (7,8). Knowledge about the contents of exosomes has contributed to the study of biomarkers in HCC, which has been useful for the prediction of disease progression and early diagnosis (9). These studies revealed the important effects of exosomes on the TME (10). Accumulating research has found that the TME is an important modulator in progression of HCC, which has enabled the increase/inhibition of exosomes as potential targets of therapeutic agents (11).
Numerous studies have investigated the role and application of microRNAs (miRNAs) in HCC. MiRNAs were found commonly in exosomes (12), and play vital roles in the pathogenesis of HCC (13). All cell types can release exosomes, which are nano-sized membrane-bound vesicles that are involved in intercellular communication (14). Exosomes can transfer biological messages between cells. These messages include miRNAs, proteins, long noncoding RNAs (lncRNAs), or DNA fragments (15). Recent studies have indicated that exosomal miRNAs might be more sensitive biomarkers compared with free miRNAs owing to their stability in exosomes (16).
LncRNA nuclear-enriched abundant transcript 1 (lncRNA-NEAT1) locates on chromosome 11 (17). Previous research has confirmed that the lncRNA NEAT1 promotes invasion and proliferation by hnRNP A2 regulation of HCC cells (18). Although an increasing number of studies have shown that NEAT1 influences multiple biological functions that play an oncogene role in HCC (17), a relationship between NEAT1 and exosome secretion or changes in exosome contents has not yet been identified. To determine whether NEAT1 contributes to exosome secretion by HCC cells, we analyzed exosome secretion by nanoparticle tracking analysis (NTA) and nanoflow cytometry (nanoFCM). We demonstrated that NEAT1 could promote exosome secretion by NEAT1-overexpressing HCC cells. Using a miRNA microarray system and reverse transcription polymerase chain reaction (RT-PCR) analysis, we found that several suppressor miRNAs changed in exosomes secreted by NEAT1-overexpressing cells. This study indicated oncogene function of NEAT1 can continue to increase with increasing NEAT1 expression levels. It will benefit for study in nude mice. This study also indicate NEAT1 might be related with TME by changing exosomal miRNAs. Taken together, our results demonstrated that NEAT1 could change exosome secretion and miRNA levels in the exosomes produced by HCC cells. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-729).
Methods
Cell culture
HCC cell lines including HuH-7, HepG2, and 293T cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China (national collection of authenticated cell cultures of China). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) supplemented with penicillin (100 U/mL) and streptomycin (100 µg/mL).
Plasmid construction and transfection
According to the instructions of manufacturer, 293T cells were transfected using Lipofectamine 2000 (Invitrogen, USA). GV641 (Shanghai Genechem Co., Ltd., China) was selected as the expression plasmid vector; the component sequence of this vector is CMV-MCS-EF1-ZsGreen1-T2A-puromycin, and the cloning site is EcoRI/XbaI. As for the transfection of miRNA mimics, HCC cells were transfected using Lipofectamine 2000 for 6 h. The target gene (NEAT1) primers, small interfering RNAs for NEAT1 (si-NEAT1), hsa-miRNAs (mimics), and cel-miR-67 [negative control (NC)] were synthesized by Shanghai Genechem Co., Ltd. (China) as follows:
NEAT1 (47382-2D)-p1: GATCTATTTCCGGTGAATTCGGAGTTAGCGACAGGGAGGGATGCGCG;
NEAT1 (47382-2D)-p2: ATGGACTCGAGTCATCTAGTTCTAATGAGTTTAGAACTCAAACTTTATTTG;
si-NEAT1: target: GCCTCCGGTCATACTAGTT, forward: GCCUCCGGUCAUACUAGUUdTdT, reverse: dTdTCGGAGGCCAGUAUGAUCAA;
hsa-miR-634 MIMAT0003304: forward: AACCAGCACCCCAACUUUGGAC, reverse: GUCCAAAGUUGGGGUGCUGGUU;
hsa-miR-638 MIMAT0003308: forward: AGGGAUCGCGGGCGGGUGGCGGCCU, reverse: AGGCCGCCACCCGCCCGCGAUCCCU;
hsa-miR-3960 MIMAT0019337: forward: GGCGGCGGCGGAGGCGGGGG, reverse: CCCCCGCCTCCGCCGCCGCC;
cel-miR-67 (NC): forward: UCACAACCUCCUAGAAAGAGUAGA, reverse: UCUACUCUUUCUAGGAGGUUGUGA.
Lentiviral infection
Vectors with recombinant NEAT1 or an empty control were cultured with 293T cells for 6 h at 37 °C and 5% CO2 for transfection. The cellular supernatant was collected 48 h after transfection for lentivirus quality examination. Next, HCC cells were infected with the viral suspension for 72 h and observed under a fluorescent microscope (micropublisher 3.3RTV, Olympus, Japan).
Exosome isolation
HCC cells were cultured for 48 hours and the culture supernatant was used to prepare the exosomes. The supernatant was collected for centrifugation (2,000 ×g for 10 min and 1,000 ×g for 30 min). Subsequently, the supernatant was collected and centrifuged at 100,000 ×g for 70 min (4 °C). The pellet was collected and resuspended in phosphate-buffered saline (PBS) as described previously (19).
NTA
Exosomes were tracked using the NanoSight NS 300 system (PARTICLE METRIX, ZetaVIEW S/N 17-310, Germany) to detect the number and size. Samples in each group were diluted 1,000–2,000 times with Dulbecco’s PBS (DPBS). Finally, the samples reached a concentration that was less than 20×108 particles/mL. Each sample was distributed to three tubes that were measured using a camera (Hitachi, HT-7700, China). According to the data, the distribution of the number and size were measured using the Stokes-Einstein equation (20).
Transmission electron microscopy
Samples were fixed using glutaraldehyde in phosphate buffer at 4 °C, and the fixed samples were sliced at a thickness of approximately 70 nm. Exosomes in the suspension droplets were placed on the copper mesh of the electron microscope (Hitachi, HT-7700, China) and then subjected to negative staining with 20 μL 2% phosphotungstic acid for 10 min. We used a H-7700 electron microscope (Hitachi, China) to analyze samples at 100 kV.
RNA extraction and RT-PCR assay
Total RNAs were extracted using TRIzol reagent (Invitrogen) and detected by nanodrop for quality. Exosomal RNAs were extracted using TRIzol reagent. For messenger RNA (mRNA), we used glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal reference, and for miRNA we used U6 as an internal reference. A High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) was used to transcribe RNA into cDNA. The reaction volume was 20 μL. RT-PCRs were analyzed in a reaction volume of 20 μL using the SYBR Green mix on an Applied Biosystems 7500. RNA expression in each group was normalized to GAPDH or U6. We used the 2−ΔΔCt method to calculate fold change. The primers were as follows:
NEAT1: forward: CTCACTCCACCCCTTCT, reverse: TCCTCCACCATTACCAA;
GAPDH: forward: GAACGGGAAGCTCACTGG, reverse: GCCTGCTTCACCACCTTCT;
Alanine-tRNA ligase, cytoplasmic (AARS): forward: CGACCATACACTGGGAAA, reverse: AGGGACTGGACGACAACA;
Heat shock protein family A member 8 (HSPA8): forward: TACAAGGGAGAGACCAAAA, reverse: GTAGCCTGACGCTGAGAGT;
U6: forward: CTCGCTTCGGCAGCACA, reverse: AACGCTTCACGAATTTGCGT;
Solute carrier family 3 member 2 (SLC3A2): forward: CGAGAAGCAGCCGATGA, reverse: TGGACAGGCCCGTGAAC;
DnaJ heat shock protein family member A1 (DNAJA1): forward: CAAACCCAATGCTACTCA, reverse: CCACCTGCTCCACCCTCT;
Heat shock protein (HSP90AA1): forward: AAGTTGAAAAGGTGGTTGTG, reverse: TGTTGAGTTGTCTCTTAGGG;
Methionyl-tRNA synthetase (MARS): forward: ACCCAATGCCCAGTTTATC, reverse: TTCTTCCACCATCTCTCCC;
Seryl-tRNA synthetase (SARS): forward: AAGCGAGCAGGGCATCAACTCC, reverse: GCCATCCTCAAAAAGCCGAAAA;
Solute carrier family 3 member 2 (SLA7A5): forward: TCTTCCTGATCGCCGTCTCC, reverse: CCACTTGGGCTTGTTTTTCC;
Transmembrane protein 109 (TMEM109): forward: CCAACAGAAGAGAGAAGC, reverse: TCCCAGACAGAGCAAAGA;
miR-6089: forward: ACACTCCAGCTGGGGGAGGCCGGGGTGGGGCG, reverse: TGGTGTCGTGGAGTCG;
miR-634: forward: ACACTCCAGCTGGGAACCAGCACCCCAACT, reverse: TGGTGTCGTGGAGTCG;
miR-3960: forward: ACACTCCAGCTGGGGGCGGCGGCGGAGG, reverse: TGGTGTCGTGGAGTCG;
miR-6127: forward: ACACTCCAGCTGGGTGAGGGAGTGGGT, reverse: TGGTGTCGTGGAGTCG;
miR-638: forward: ACACTCCAGCTGGGAGGGATCGCGGGCGGGTGG, reverse: TGGTGTCGTGGAGTCG.
Cell proliferation assay
Cells were seeded in 96-well plates with a density of 2,000 per well. We used a Cell Counting Kit-8 (CCK-8) assay to analyze cell viability at 0, 24, 48, and 72 h after seeding. An automatic microplate reader (Infinite M1000, TECAN, Switzerland) was used to detect absorbance at 450 nm.
Invasiveness analysis
The invasive function was assessed with a Transwell assay (Millipore, Billerica, MA, USA). A total of 1×104 HCC cells were suspended in DMEM without serum. Next, the cells were seeded in the upper chamber. Matrigel (BD Biosciences, USA) (200 mg/mL, 100 mL) was used to cover the membrane. To produce a chemoattractant environment, the lower chamber contained FBS (final concentration: 15%). After culturing for 48 h, we cleaned the cells in the upper chamber. Invasive cells were fixed with methanol and stained with 0.1% crystal violet. For calculation, cells in the lower chamber were detached by trypsin after cleaning cells in the upper chamber. The number of cells was counted using a hemocytometer (Z359629-1EA, Merck, China).
FCM analysis
For apoptosis analysis, we used propidium iodide (PI) and annexin V-FITC (BestBio, Shanghai, China) to incubate with HCC cells for 15 minutes at room temperature. FCM was performed using a FACSCalibur device (BD Biosciences, USA), and the data were analyzed using Cell Quest software (BD Biosciences, USA). For exosome FCM analysis, the exosomes were diluted and incubated with CD9-FITC, CD81-FITC and IgG-FITC (BD Biosciences) for 30 minutes at 37 °C. FCM was performed using a NanoFCM (N30E).
RNA sequencing and data analysis
The integrity of RNA was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). A RNA integrity number of samples (RIN) ≥7 was used for analysis. Libraries were constructed using the stranded mRNA prep kit (Illumina, USA), and were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten). Next, 125 bp/150 bp paired-end reads were generated. Feature Extraction software (version 10.7.1.1, Agilent Technologies) was used for microarray analysis. Raw data were normalized using the quantile algorithm. Probes that had flags of “Detected” in any one of two conditions were selected for further data analysis. Changed miRNAs were then identified through the fold change and P value using t-tests. A fold change ≥2 and P value <0.05 were set for up- and down-regulated genes. Target miRNAs were predicted using two databases (miRDB, miRWalk). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were applied to determine the roles of target genes. Hierarchical clustering was performed to display the different miRNA expression patterns.
Statistical analysis
Analyses were performed using Prism 8.0 software (USA). Analyses included the Student’s t-test and two-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons. The data graph was presented using Prism GraphPad 8.0, and significant differences between each group were marked using *, P<0.05, **, P<0.01, ***, P<0.001, or ****, P<0.0001.
Results
NEAT1-overexpression promoted the invasion and proliferation of HCC cells
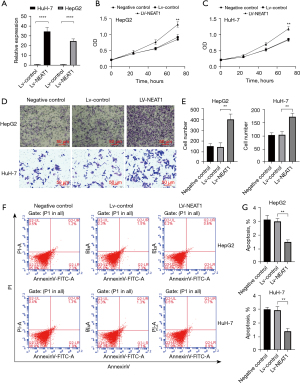
The efficacy of the NEAT1 overexpression was measured in HCC cells (approximately 30 times for HepG2 cells and 20 times for HuH-7 cells; Figure 1A). Previous studies assessed functions of NEAT1 knockdown cells. The aim of our research was to confirm whether the oncogene function could increase with increasing NEAT1 expression levels, as NEAT1 expression levels are naturally high in HCC cell lines and tissues. Proliferation was assessed by CCK-8 assays, and these experiments showed that overexpression of NEAT1 significantly promoted the proliferation of HCC cells compared with the Lv-control and NC groups (P<0.01; Figure 1B,1C). We also analyzed the effect of Lv-NEAT1 on invasiveness using Transwell assays. Compared with the Lv-control and NC groups, NEAT1 overexpression promoted invasiveness, which was observed using crystal violet staining (Figure 1D). Markedly more cells detached from the lower chamber filters in the Lv-NEAT1 group compared with the Lv-control and NC groups (P<0.01; Figure 1E). Apoptosis was assessed by FCM, which indicated that overexpression of NEAT1 considerably decreased the apoptosis of HCC cells compared with the Lv-control and NC groups (P<0.01; Figure 1F,1G).
Overexpression of NEAT1 promoted exosome secretion by HCC cells
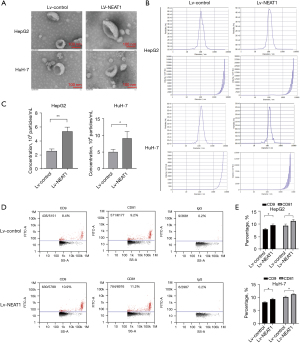
A recent study showed that lncRNAs can regulate abnormal exosome-related gene expression, which results in abnormal exosome secretion by HCC cells (20). We used transmission electron microscopy to detect the purity of exosomes in each group. As shown in Figure 2A, for each group, the exosomes were double membrane and 100–150 nm in diameter. NTA indicated that the sizes of the released exosomes were approximately 150 nm (Figure 2B). By combining the particle number and volume (Figure 2B), we found that exosomes were secreted in greater quantities by the overexpression of NEAT1 HCC cells than by the Lv-control group (P<0.05) (Figure 2C). To explore surface molecular differences of exosomes, we detected the markers (CD81 and CD9) of the Lv-NEAT1 group compared with those of the Lv-control group. The data indicated that NEAT1 overexpression increased exosome secretion (Figure 2D,2E). Exosomes released by NEAT1 overexpression cells contained CD81 and CD9 (Figure 2D). These results indicated that NEAT1 could affect the secretion of exosomes in HCC cells.
Overexpression of NEAT1 caused global gene expression changes in HCC cells
The data showed that the expression levels of 1,158 genes were changed in the NEAT1-overexpressing cells (including 362 up-regulated and 796 down-regulated genes, fold change ≥2.0 and P value <0.05) (Figure 3A). GO analysis indicated that the changed genes were involved in biological and molecular functions, including extracellular matrix changes (Figure 3B). The top 20 variations after the overexpression of NEAT1 were presented in Figure 3B, which were associated with biological and molecular functions. Also, KEGG pathway analysis identified variations in the MAPK signaling pathway, apoptosis, p53 signaling pathway, and the HIF-1 signaling pathway after overexpression of NEAT1 (Figure 3C).
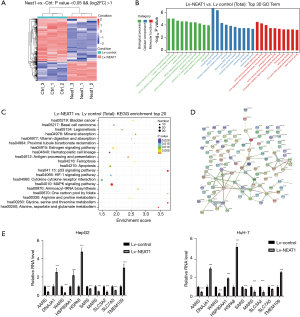
Based on the bioinformatics analysis data as well as the results showing that NEAT1 overexpression promoted HCC cell invasion, proliferation, and exosome secretion, we selected genes that are involved in cellular components relevant to exosome synthesis and secretion. We found that these genes were associated with each other. We drew a protein-protein interaction network of these genes based on the bioinformatics analysis (Figure 3D). Using RT-PCR assays, we confirmed the upregulation of HSPA8, DNAJA1, HSP90AA1, and TMEM109, and the downregulation of AARS, SARS, MARS, SLC3A2, and SLC7A5 in NEAT1-overexpressing HCC cells (Figure 3E). We found that the PCR results matched the protein-protein interaction network of these exosome-related genes.
Overexpression of NEAT1 altered global miRNA levels in HCC cells
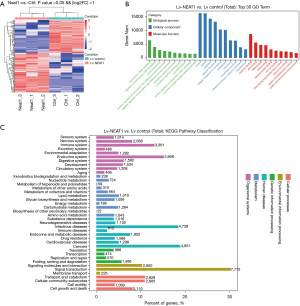
MiRNAs play a vital role in the interaction between NEAT1 and HCC pathogenesis. In addition, it is possible that NEAT1-related miRNAs participate in exosome synthesis, secretion regulation, and even exosome-containing changes. We used gene sequencing to analyze miRNA level changes in HCC cells 72 h after Lv-NEAT1 infection. Overall, 169 miRNAs were differentially expressed in NEAT1-overexpressing cells (including 92 upregulated and 77 downregulated miRNAs, fold change >2.0 and P value <0.05) (Figure 4A). Combined with the miRNA-mRNA interaction analysis, GO analysis showed that the changed miRNA-related mRNAs were involved in biological and molecular functions, which were related to HCC pathogenesis, including exosome and extracellular matrix changes (Figure 4B). KEGG pathway analysis identified the classification and target mRNA number of variation miRNA levels after NEAT1 overexpression (Figure 4C). The data show that miRNAs regulated by NEAT1 reacted to a large number of mRNAs, which significantly contributed to cancer pathogenesis (Figure 4C).
Overexpression of NEAT1 changed miRNAs carried by exosomes in HCC cells
MiRNAs carried by exosomes (exosomal miRNAs) secreted by HCC cells play a vital role in cellular communication, which might contribute to tumor growth, metastasis, angiogenesis, and immunologic escape. NEAT1 changes exosome secretion and induces global miRNA level changes in HCC cells, and it is possible that NEAT1 also changes the miRNA content in exosomes. We used a miRNA microarray system to analyze the miRNA level changes in exosomes secreted by HCC cells 72 h after Lv-NEAT1 infection, in order to identify the miRNA changes carried by the exosomes. The data showed that five miRNA levels were different in exosomes secreted by NEAT1-overexpressing cells. All altered miRNA levels were downregulated (including one miRNA, fold change >5.0 and P value <0.05 and four miRNAs, fold change >2.0 and P value <0.05) (Figure 5A).
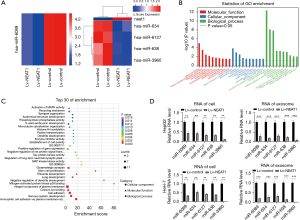
Combined with the miRNA-mRNA interaction analysis, GO analysis showed that the changed miRNA-related mRNAs were involved in biological and molecular functions relevant to the pathogenesis of HCC (Figure 5B). KEGG pathway analysis identified variations in MAPK activity that are associated with NEAT1 and HCC progression, and identified a close association between miRNA and mRNA changes caused by NEAT1 in our bioinformatics analysis and previous work (Figure 5C). We confirmed the downregulation of miR-6089, miR-634, miR-638, miR-6127, and miR-3960 in NEAT1-overexpressing HCC cells by RT-PCR assays (Figure 5D).
In addition to bioinformatics analysis data, we also found that most altered miRNAs in exosomes, such as miR-6089, miR-634, miR-638 and miR-3960, played suppressor gene functions in HCC, which has been shown in other studies. It will be interesting to confirm whether NEAT1 downregulated these suppressor miRNAs, which are transported between cells carried by exosomes in HCC. Using RT-PCR assays, we also confirmed that miR-6089, miR-634, miR-638, miR-6127, and miR-3960 were downregulated in exosomes secreted by NEAT1-overexpressing HCC cells (Figure 5D).
MiR-634, miR-638, and miR-3960 reverse proliferation and invasion function in NEAT1-overexpressing HCC cells
We observed the alteration of several exosomal miRNAs in NEAT1-overexpressing HCC cells. We proposed the hypothesis that NEAT1 promotes cell progression through regulation of exosomal RNAs. We used miRNA mimics of downregulated exosomal RNAs to transfer NEAT1-overexpressing HCC cells and NEAT1-knockdown HCC cells. We found that transfection of miR-634, miR-638, and miR-3960 reversed the enhanced proliferation function in NEAT1-overexpressing HCC cells (P<0.01, Figure 6A). However, the reversion was not observed in NEAT1-knockdown HCC cells because transfection of these miRNA mimics did not significantly reduce proliferation in NEAT1-knockdown HCC cells (Figure 6A).
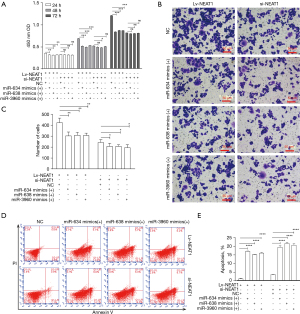
Transfection of miR-634, miR-638, and miR-3960 reduced invasion in NEAT1-overexpressing HCC cells and NEAT1-knockdown HCC cells (P<0.05; Figure 6B,6C). In NEAT1-knockdown HCC cells, the extent of inhibition might be relenting compared NEAT1-overexpressing HCC cells group (Figure 6B,6C). Transfection of miR-634, miR-638, and miR-3960 markedly increased the apoptosis rate in HCC cells of Lv-NEAT1 and si-NEAT1 groups (P<0.0001; Figure 6D,6E). As for the change of apoptosis, there was no difference in the extent of increase between NEAT1-overexpressing HCC cells and NEAT1-knockdown HCC cells (Figure 6D,6E).
Discussion
Our previous study determined that lncRNA NEAT1 was an oncogene. When NEAT1 was downregulated, the invasion and proliferation of HCC cells were inhibited, as was tumor growth in xenograft models. These effects were correlated with hnRNP A2, which is regulated by NEAT1 (18). A number of studies have confirmed the oncogene function of NEAT1 and its specific molecular mechanism, including its function through lncRNA-miRNA interactions in cells [miR-124-3p (21), miR-204 (22), miR-296-5p (23), miR-22-3p (24), miR-613 (25), miR-485 (26)], its function through the lncRNA-RBP complex [NEAT1-U2AF65 (18), NEAT1-PSPC1 (27)], and its function through gene regulation [TUG1 (28), ATGL (21), TGF-β1 (29), STAT3 (26)]. Most of these studies confirmed the function and mechanism associated with intracellular regulation using NEAT1 knockdown in HCC cell lines. Hence, there are two problems to be solved according to the current study of NEAT1. The first problem is whether the oncogene function of NEAT1 can continue to increase with increasing NEAT1 expression levels, and the second issue relates to the identification of the different mechanisms.
NEAT1 has a high expression level in HCC cell lines and tissues (30), which makes it possible that the basic expression level of NEAT1 could perform an oncogene function in HCC and that overexpression of NEAT1 might not increase the intensity of its oncogene function. In previous work on NEAT1, one study used NEAT1-overexpressing cells instead of NEAT1-knockdown cells to explore the metastatic function and mechanism of HCC in vitro and in vivo under hypoxia, as decreasing the growth and metastatic function of NEAT1-knockdown cells did not demonstrate positive findings in a metastasis model (31). Compared with the NEAT1 knockdown HCC cell model, the NEAT1 overexpression cell model has advantages in tumor growth and metastasis studies in vivo. NEAT1-overexpressing cells also made it possible to explore extracellular matrix changes that might be associated with NEAT1 overexpression. Our study confirmed that lentivirus transfection was a reliable method for NEAT1 overexpression in HCC cell lines and that the oncogene function of NEAT1 was not limited to its basic level of expression in HCC cell lines (Figure 1). Our study also revealed alterations in global gene expression patterns and miRNA levels in HCC cells, which might be associated with cancer pathogenesis in NEAT1-overexpressing HCC cells (Figures 3,4). These data might support further NEAT1 overexpression studies on metastasis, drug resistance, hypoxia, angiogenesis, and extracellular matrix changes in vivo, considering the various functions of lncRNAs (12).
A recent study showed that lncRNAs regulate a large number of mRNAs and miRNAs in HCC, which suggests that lncRNAs affect exosome secretion (32,33). Recently, several studies have suggested that lncRNAs can regulate exosome secretion, such as lncRNA HULC/miR-372-3p, which plays a significant role in exosome secretion. LncRNA HOTAIR was found to promote exosome secretion by inducing the phosphorylation of SNAP23, which could be a mammalian target of rapamycin (mTOR) signaling (20,34). LncRNA NEAT1 has various significant biological functions associated with RNA-binding proteins (RBPs), which form functional ribonucleoprotein (RNP) complexes. RBP has encouraged most NEAT1-related studies to focus on intracellular gene regulation or even paraspeckles (17), and few studies have focused on the association between NEAT1 and exosomal miRNA.
In this study, we confirmed that NEAT1 overexpression promoted exosome secretion by NTA and nanoFCM (Figure 3). Combining the bioinformatics analyses of NEAT1-overexpressing cells, we found that a large number of altered genes were involved in exosome secretion-related cellular components, including the plasma membrane, cellular exosomes, and PKM2-related pathways (Figures 3,4). Using RT-PCR assays, we confirmed the presence of HSPA8, a traditional exosome marker (35) with high expression in NEAT1-overexpressing cells (Figure 3D). HSPA8 also affects the expression of CD38, which is expressed on the surface of secreted exosomes (36). Based on the protein-protein interaction analysis, the expression of MARS, WARS, AARS, and SLC proteins might change due to HSPA8 alterations (Figure 3D). These proteins are also associated with secretion and are contained in exosomes (37-39). Our RT-PCR data confirmed that the expression of exosome- or HSPA8-related genes, including DNAJA1, HSP90AA1, TMEM109, AARS, SARS, MARS, SLC3A2, and SLC7A5, was altered in NEAT1-overexpressing cells. Up- or down-regulated regulation of these genes was consistent with the bioinformatics analysis and the protein-protein interaction network, which indicates that NEAT1 plays a vital role in exosome-related gene expression.
The phenomenon of increased exosome secretion in HCC is common; however, the molecular mechanisms have not been elucidated (20). Exosomes transfer effector messages between cells, including miRNAs, proteins, or DNA fragments (15,40). Exosomes have been studied for many years, but the biological roles of exosomal miRNAs are only just beginning to be understood in HCC (16). Exosomal miRNAs contribute to HCC progression and metastasis signaling networks, including cell metastasis, proliferation, immune escape, and interactions with the TME (16,32,41). Recent studies have investigated several types of exosomes and confirmed their biological function in HCC, such as exosomes containing miR-122 secreted by Huh7 cells that inhibit growth and proliferation by targeting CAT1 (42), exosomes containing miR-142 secreted by macrophages that inhibit proliferation by targeting STMN1 (43), exosomes containing miR-320a secreted by CAFs that inhibit migration and progression by targeting the MAPK pathway (44), and exosomes containing miR-1247-3p secreted by LM3 cells that promote the conversion of normal fibroblasts into CAFs and accelerate the lung metastasis of HCC by targeting B4GALT3 (32).
Exosomal miRNAs might be more sensitive biomarkers compared with free miRNAs owing to their stability in exosomes (16). This study focused on biological roles and function of exosomal miRNAs regulated by NEAT1 in HCC. The mechanism of NEAT1-exosomal miRNAs-target mRNAs could provide potential target for HCC diagnosis and therapy. Exosomes can play role of carriers for gene target therapy. Thus, advance study can try to use exosomal miRNAs as nucleic acid and exosomes as carrier for HCC target therapy study. Considering the exosome secretion-related function of NEAT1, the global alteration of miRNA levels in NEAT1-overexpressing cells, and the fact that the target mRNAs of altered miRNAs are involved in HCC pathogenesis (Figure 4), we analyzed exosomal miRNAs in the NEAT1-overexpressing and control groups using a miRNA microarray system, and confirmed an alteration of exosomal miRNAs by RT-PCR. Interestingly, we found that in NEAT1-overexpressing HCC cells, most of the exosomal miRNAs were downregulated (Figure 5A), and they were previously shown to have tumor suppressor functions in HCC, such as miR-6089 (45), miR-634 (46), miR-638 (47-49), and miR-3960 (50). Our data also indicated that exosomal miRNAs were also downregulated in NEAT1-overexpressing HCC cells (Figure 5D). These data imply that increasing NEAT1 expression downregulates several suppressor miRNAs both in cells and also in exosomes secreted by HCC cells. Therefore, confirming the suppressor function of these exosomal miRNAs will be meaningful for exosome-mediated miRNA transfer for HCC therapy, because compared with free miRNAs, exosomal miRNAs have much greater stability and possess important biological properties that ensure long-term persistence (16,51).
To determine whether NEAT1 promotes HCC progression by regulation of exosomal miRNA, we used these changed exosomal miRNA mimics to transfer NEAT1-overexpressing HCC cells and NEAT1-knockdown HCC cells and detect cell function. Transfection of miR-634, miR-638, and miR-3960 reversed the enhancement of proliferation and invasion by NEAT1 in NEAT1-overexpressing HCC cells (Figure 6). However, this reversion phenomenon was not observed or relenting in NEAT1-knockdown HCC cells (Figure 6). Based on the fact that NEAT1 significantly down-regulated miR-634, miR-638, and miR-3960 in exosomes, these data suggest that the down-regulation or transmission reduction of miR-634, miR-638, and miR-3960 in exosomes might contribute to cell invasion and proliferation in HCC cells with a high level of NEAT1 expression. It also indicated that the suppressor functions of miR-634, miR-638, and miR-3960 are reliant on NEAT1 expression. However, increased apoptosis by miR-634, miR-638, and miR-3960 mimics might have no association with NEAT1 expression, as transfection of these miRNAs the increased apoptosis rate and there is no difference in the extent of increase between NEAT1-overexpressing and NEAT1-knockdown HCC cells (Figure 6). Bioinformatics data will not be used in clinic as clinicopathological and prognostic biomarker directly. We need to screen out potential data by cell lines and nude mice verification. In this study, we focused on NEAT1-miRNAs function. Our Bioinformatics data shown more than 100 miRNAs and target mRNAs change. However, not all miRNAs and target mRNAs can be biomarker. Thus, stability, function and relationship with NEAT1 need to be verification. This study using exosomal miRNA to screen out potential miRNAs which might be stable. We also use miRNA mimics to verified function of exosomal miRNA to make sure these miRNAs as tumor suppressor gene. To explore if these miRNAs’ functions are related with NEAT1, we used these changed exosomal miRNA mimics to transfer NEAT1-overexpressing HCC cells and NEAT1-knockdown HCC cells and found proliferation and invasion reversion phenomenon was not observed or relenting in NEAT1-knockdown HCC cells. It indicated function of NEAT1 might be related with exosomal miRNA change. The same method will be used in target mRNA in vivo, vitro and clinical samples. Finally, clinicopathological and prognostic biomarker might be found combined these data. Taken together, the results of our study demonstrated that NEAT1 promotes cell invasion and proliferation through downregulation of miR-634, miR-638, and miR-3960 in exosomes.
Conclusions
This study indicates that lncRNA NEAT1-overexpression promotes proliferation and invasion, and inhibits apoptosis in HCC cells. Overexpression of NEAT1 also promotes exosome secretion by HCC cells. This effect might be correlated with altered gene expression related to exosome synthesis and secretion, which is also regulated by NEAT1. Increasing NEAT1 expression downregulated several suppressor miRNAs both in cells and exosomes secreted by HCC cells. Furthermore, NEAT1 promoted cell proliferation and invasion via downregulation of miR-634, miR-638, and miR-3960 in exosomes. The suppressor function of miR-634, miR-638, and miR-3960 were found to be reliant on NEAT1 expression.
Acknowledgments
This study was performed in the Biomedical Research Center of The Calmette Affiliated Hospital of Kunming Medical University, The First Hospital of Kunming. We would like to thank the Biomedical Research Center of the First Hospital of Kunming for their molecular experiment support. We thank Shanghai OE Biotech Inc. (Shanghai, China) for high-throughput sequencing service and bioinformatics support.
Funding: These studies were supported by grants from the Medical Academic Leaders Training Program of Yunnan Provincial Health and the Family Planning Commission (No. D-201616, to SZ), and the Kunming Science and Technology Bureau (No. 2019-1-C-25318000002252, to YM).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-729
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-729
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-729). All authors report that they received high-throughput sequencing service and bioinformatics support from Shanghai OE Biotech Inc. (Shanghai, China). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol 2015;13:2140-51. [Crossref] [PubMed]
- Zhu Q, Li N, Zeng X, et al. Hepatocellular carcinoma in a large medical center of China over a 10-year period: evolving therapeutic option and improving survival. Oncotarget 2015;6:4440-50. [Crossref] [PubMed]
- De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer 2017;17:457-74. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- El-Magd MA, Mohamed Y, El-Shetry ES, et al. Melatonin maximizes the therapeutic potential of non-preconditioned MSCs in a DEN-induced rat model of HCC. Biomed Pharmacother 2019;114:108732. [Crossref] [PubMed]
- Kalra H, Drummen GP, Mathivanan S. Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci 2016;17:170. [Crossref] [PubMed]
- Fan Q, Yang L, Zhang X, et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett 2018;414:107-15. [Crossref] [PubMed]
- Alzahrani FA, El-Magd MA, Abdelfattah-Hassan A, et al. Potential effect of exosomes derived from cancer stem cells and MSCs on progression of DEN-induced HCC in rats. Stem Cells Int 2018;2018:8058979. [Crossref] [PubMed]
- Liu H, Li B. The functional role of exosome in hepatocellular carcinoma. J Cancer Res Clin Oncol 2018;144:2085-95. [Crossref] [PubMed]
- O'Brien K, Rani S, Corcoran C, et al. Exosomes from triple-negative breast cancer cells can transfer phenotypic traits representing their cells of origin to secondary cells. Eur J Cancer 2013;49:1845-59. [Crossref] [PubMed]
- Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 2011;17:320-9. [Crossref] [PubMed]
- Klingenberg M, Matsuda A, Diederichs S, et al. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol 2017;67:603-18. [Crossref] [PubMed]
- Yuan J, Li Y, Liao J, et al. MicroRNA-7 inhibits hepatocellular carcinoma cell invasion and metastasis by regulating Atg5-mediated autophagy. Transl Cancer Res 2020;9:3965-72. [Crossref]
- Guo S, Chen W, Luo Y, et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol 2015;8:5395-402. [PubMed]
- Kosaka N. Decoding the secret of cancer by means of extracellular vesicles. J Clin Med 2016;5:22. [Crossref] [PubMed]
- Li S, Yao J, Xie M, et al. Exosomal miRNAs in hepatocellular carcinoma development and clinical responses. J Hematol Oncol 2018;11:54. [Crossref] [PubMed]
- Hirose T, Yamazaki T, Nakagawa S. Molecular anatomy of the architectural NEAT1 noncoding RNA: the domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip Rev RNA 2019;10:e1545. [Crossref] [PubMed]
- Mang Y, Li L, Ran J, et al. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther 2017;10:1003-16. [Crossref] [PubMed]
- Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3:Unit 3.22.
- Yang L, Peng X, Li Y, et al. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer 2019;18:78. [Crossref] [PubMed]
- Liu X, Liang Y, Song R, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer 2018;17:90. [Crossref] [PubMed]
- Li X, Zhou Y, Yang L, et al. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol 2020;235:3402-13. [Crossref] [PubMed]
- Li Y, Ding X, Xiu S, et al. LncRNA NEAT1 promotes proliferation, migration and invasion via regulating miR-296-5p/CNN2 axis in hepatocellular carcinoma cells. Onco Targets Ther 2019;12:9887-97. [Crossref] [PubMed]
- Zhou X, Wang X, Zhou Y, et al. Long noncoding RNA NEAT1 promotes cell proliferation and invasion and suppresses apoptosis in hepatocellular carcinoma by regulating miRNA-22-3p/akt2 in vitro and in vivo. Onco Targets Ther 2019;12:8991-9004. [Crossref] [PubMed]
- Wang Z, Zou Q, Song M, et al. NEAT1 promotes cell proliferation and invasion in hepatocellular carcinoma by negative regulating miR-613 expression. Biomed Pharmacother 2017;94:612-8. [Crossref] [PubMed]
- Zhang XN, Zhou J, Lu XJ. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J Cell Physiol 2018;233:6733-41. [Crossref] [PubMed]
- Jen HW, Gu DL, Lang YD, et al. PSPC1 potentiates IGF1R expression to augment cell adhesion and motility. Cells 2020;9:1490. [Crossref] [PubMed]
- Mohyeldeen M, Ibrahim S, Shaker O, et al. Serum expression and diagnostic potential of long non-coding RNAs NEAT1 and TUG1 in viral hepatitis C and viral hepatitis C-associated hepatocellular carcinoma. Clin Biochem 2020;84:38-44. [Crossref] [PubMed]
- Tu J, Zhao Z, Xu M, et al. NEAT1 upregulates TGF-β1 to induce hepatocellular carcinoma progression by sponging hsa-mir-139-5p. J Cell Physiol 2018;233:8578-87. [Crossref] [PubMed]
- Bu FT, Wang A, Zhu Y, et al. LncRNA NEAT1: Shedding light on mechanisms and opportunities in liver diseases. Liver Int 2020;40:2612-26. [Crossref] [PubMed]
- Zheng X, Zhang Y, Liu Y, et al. HIF-2α activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition. J Cell Biochem 2018;119:3247-56. [Crossref] [PubMed]
- Fang T, Lv H, Lv G, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun 2018;9:191. [Crossref] [PubMed]
- Sun Z, Yang S, Zhou Q, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer 2018;17:82. [Crossref] [PubMed]
- Cao SQ, Zheng H, Sun BC, et al. Long non-coding RNA highly up-regulated in liver cancer promotes exosome secretion. World J Gastroenterol 2019;25:5283-99. [Crossref] [PubMed]
- Hoshino A, Kim HS, Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020;182:1044-1061.e18. [Crossref] [PubMed]
- Zumaquero E, Muñoz P, Cobo M, et al. Exosomes from human lymphoblastoid B cells express enzymatically active CD38 that is associated with signaling complexes containing CD81, Hsc-70 and Lyn. Exp Cell Res 2010;316:2692-706. [Crossref] [PubMed]
- Chen M, Li Y, Lv H, et al. Quantitative proteomics and reverse engineer analysis identified plasma exosome derived protein markers related to osteoporosis. J Proteomics 2020;228:103940. [Crossref] [PubMed]
- Console L, Scalise M, Indiveri C. Exosomes in inflammation and role as biomarkers. Clin Chim Acta 2019;488:165-71. [Crossref] [PubMed]
- Seyfizadeh N, Seyfizadeh N, Rahbarghazi R, et al. Isolation and characterization of human amniotic fluid and SH-SY5Y/ BE(2)-M17 cell derived exosomes. Acta Neurobiol Exp (Wars) 2019;79:261-9. [Crossref] [PubMed]
- Zhang L, Valencia CA, Dong B, et al. Transfer of microRNAs by extracellular membrane microvesicles: a nascent crosstalk model in tumor pathogenesis, especially tumor cell-microenvironment interactions. J Hematol Oncol 2015;8:14. [Crossref] [PubMed]
- Wu Q, Zhou L, Lv D, et al. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol 2019;12:53. [Crossref] [PubMed]
- Basu S, Bhattacharyya SN. Insulin-like growth factor-1 prevents miR-122 production in neighbouring cells to curtail its intercellular transfer to ensure proliferation of human hepatoma cells. Nucleic Acids Res 2014;42:7170-85. [Crossref] [PubMed]
- Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol 2013;191:6250-60. [Crossref] [PubMed]
- Zhang Z, Li X, Sun W, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett 2017;397:33-42. [Crossref] [PubMed]
- Liu L, Ning Y, Yi J, et al. miR-6089/MYH9/β-catenin/c-Jun negative feedback loop inhibits ovarian cancer carcinogenesis and progression. Biomed Pharmacother 2020;125:109865. [Crossref] [PubMed]
- Zhang CZ, Cao Y, Fu J, et al. miR-634 exhibits anti-tumor activities toward hepatocellular carcinoma via Rab1A and DHX33. Mol Oncol 2016;10:1532-41. [Crossref] [PubMed]
- Shi M, Jiang Y, Yang L, et al. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem 2018;119:4711-6. [Crossref] [PubMed]
- Yang J, Li B, Zhao S, et al. Exosomal miR-638 Inhibits Hepatocellular Carcinoma Progression by Targeting SP1. Onco Targets Ther 2020;13:6709-20. [Crossref] [PubMed]
- Zhang Y, Zhang D, Jiang J, et al. Loss of miR-638 promotes invasion and epithelial-mesenchymal transition by targeting SOX2 in hepatocellular carcinoma. Oncol Rep 2017;37:323-32. [Crossref] [PubMed]
- Mo F, Luo Y, Fan D, et al. Integrated Analysis of mRNA-seq and miRNA-seq to Identify c-MYC, YAP1 and miR-3960 as Major Players in the Anticancer Effects of Caffeic Acid Phenethyl Ester in Human Small Cell Lung Cancer Cell Line. Curr Gene Ther 2020;20:15-24. [Crossref] [PubMed]
- Li C, Xu X. Biological functions and clinical applications of exosomal non-coding RNAs in hepatocellular carcinoma. Cell Mol Life Sci 2019;76:4203-19. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


