
Bioinformatics analysis identifying FBXO45 gene as a potential oncogene in esophageal cancer
Introduction
Esophageal cancer (ESCA) is a common gastrointestinal tumor, with approximately 300,000 patient deaths from ESCA every year worldwide (1,2). Surgery, radiotherapy, and chemotherapy are the main methods for the treatment of ESCA. However, the prognosis of patients with ESCA remains poor. It has been reported that patients with ESCA have the sixth worst prognosis owing to its aggressiveness and poor survival (3). Thus, identifying new and effective therapeutic targets for ESCA patients is crucial.
Previous studies have reported that the prognosis of patients with ESCA is associated with the tumor microenvironment (TME), specifically the tumor immune microenvironment (TIME) (4). The immunosuppressive microenvironment of ESCA is one of the main reasons for the poor prognosis of patients. Identifying the oncogenes that affect the TIME and developing target therapy may be a potential treatment for ESCA patients. TME is a vital space for the metabolism of tumor cells. Nowadays, numerous studies have shown that TME could mainly promote the growth, proliferation, migration, and metastasis of EC. The main components of TME are a variety of adaptive and innate immune cells, fibroblasts, adipocytes, endothelial cells, and extracellular matrix (ECM) components, which have been extensively studied in a variety of tumors (5-7). TME is not a fixed tumor survival environment, but a dynamic environment that is constantly remodeled by tumors and tumor-related cells to adapt to the survival of tumor cells. As the oncogenic mutation of tumor precursor cells continues to accumulate, the precancerous lesions further develop into cancer. Cancer tissues continue to deteriorate by interacting with TME and eventually lead to the occurrence of metastasis.
F-box protein 45 (FBXO45), a substrate-recognition subunit of E3 ligases, is a member of the F-box protein family. FBXO45 has been shown to be involved in the progression of many tumors. The biological functions of FBXO45 lies in the regulation of proliferation, apoptosis, cell cycle, motility and metastasis. For example, according to The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus data, FBXO45 is highly expressed in squamous-cell lung carcinoma (SCLC) tissues compared with normal lung tissues. High FBXO45 expression is correlated with poor survival of SCLC (8). It was also illustrated that high expression of FBXO45 was correlated with poor survival in patients with SCLC by obtaining data from the Kaplan–Meier plotter website and the TCGA database (8). FBXO45 expression was higher in gastric cancer tissues than in normal gastric tissues (9). Moreover, Dahlem et al. (10) proved that overexpression of insulin-like growth factor 2 (IGF2) mRNA-binding protein IMP2 (IGF2BP2) existed and was associated with poor outcomes in pancreatic cancer patients by investigating publicly available datasets FBXO45 is positively associated with IMP2 expression, indicating that FBXO45 might play a potential oncogenic role in pancreatic cancer progression.
However, the role of FBXO45 in ESCA and its correlation with the TIME remains unclear. In this study, we analyzed and verified the role of FBXO45 in ESCA. The correlation between FBXO45 expression and immune cell infiltration were also assessed. Patients with high expression of FBXO45 may have a tumor immunosuppressive microenvironment. Targeting FBXO45 may be a potential treatment of ESCA. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-662).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Patients and specimen characteristics
The RNA (Ribonucleic Acid) expression profiles from TCGA database and the Genotype-Tissue Expression (GTEx) project were downloaded from UCSC XENA website (https://xenabrowser.net/datapages/). The RNAseq data were normalized into log2(TPM+0.001). There were 204 cases of ESCA in TCGA, including one case of T0, 42 cases of T1, 44 cases of T2, 92 cases of T3, five cases of T4, and the remaining 20 cases were unclear (American Joint Committee on Cancer, AJCC 7th). Also, there were 86 cases of N0, 72 cases of N1, 14 cases of N2, 12 cases of N3, and the remaining 20 cases were unclear (AJCC 7th). The mean age of the patients was 63±11.9 (range, 27–90) years.
Assay methods
Online analysis
The pan-cancer expression of FBXO45 was assessed using the TIMER2 database (http://timer.comp-genomics.org/). The expression of FBXO45 in different tumor stages and histology subtypes were evaluated using the UALCAN database (http://ualcan.path.uab.edu/). The protein-protein interaction (PPI) network was constructed using the STRING database (https://string-db.org/).
Enrichment analysis
The correlation analysis between FBXO45 and other genes was conducted using ESCA data from TCGA cohort. The Pearson correlation coefficient was calculated. The top 300 genes most positively associated with FBXO45 were screened to perform enrichment analysis. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using R package “clusterProfiler” ((http://bioconductor.riken.jp/packages/3.10/bioc/html/clusterProfiler.html) ), according to the following parameters: P value cutoff =1, and q-value cutoff =1. The top 20 terms of each analysis were displayed.
Immune infiltration analysis
Immune cell infiltration data were downloaded from the ImmuCellAI database (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/). The correlation between FBXO45 expression and the infiltration level of 24 immune cells were analyzed. In addition, the different infiltration levels of the 24 immune cells were compared in high and low FBXO45 expression groups in ESCA.
Immunohistochemistry and immunofluorescence analysis
The immunohistochemistry and immunofluorescence data of FBXO45 in ESCA were downloaded from the Human Protein Atlas website (https://www.proteinatlas.org/).
Study design
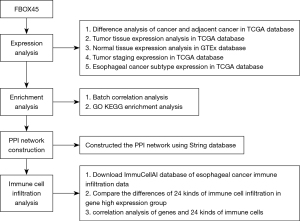
The expression analysis of FBOX45 were lies in difference analysis of cancer and adjacent cancer in TCGA database, tumor tissue expression analysis in TCGA database, normal tissue expression analysis in GTEx database, tumor staging expression in TCGA database, esophageal cancer subtype expression in TCGA database. Then we analysis the enrichment, PPI network and immune cell infiltration of FBOX45 (Figure 1).
Statistical analysis
Data were presented as mean ± standard deviation (SD). Differences between groups were analyzed using the Student’s t-test. Statistical analyses were performed using R 4.1.1 (https://www.r-project.org/). P<0.05 (two-tailed) was considered statistically significant.
Results
FBXO45 expression analysis
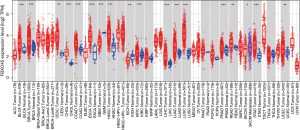
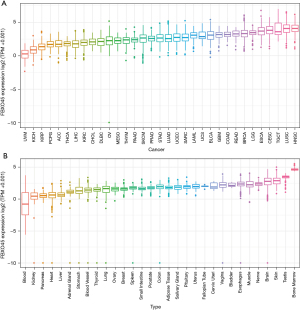
We first assessed the pan-cancer expression of FBXO45 using the TIMER2 database. The results showed that FBXO45 was over-expressed in 15 tumor types, including ESCA (Figure 2). In addition, for FBXO45 expression only in tumor tissues in TCGA cohort, FBXO45 expression was highest in HNSC (head neck squamous cell carcinoma) and lowest in UVM (uveal melanoma) (Figure 3A). For FBXO45 expression in normal tissues from the GTEx cohort, FBXO45 expression was highest in bone marrow and lowest in blood (Figure 3B).
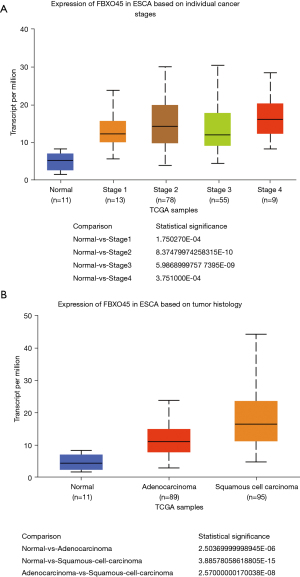
We further explored the correlation between FBXO45 and the clinical features of ESCA patients using the UALCAN database. The results revealed that FBXO45 expression was relative higher in worse tumor stages (Figure 4A). In addition, FBXO45 was highly expressed in esophageal squamous cell carcinoma (Figure 4B).
Enrichment analysis of FBXO45
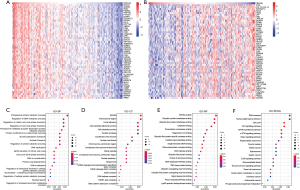
Next, we conducted the enrichment analysis using R package “clusterProfiler” to predict the function and pathways that FBXO45 might be involved in. The correlation analysis was performed and genes most correlated with FBXO45 in ESCA were identified (Figure 5A,5B). The top 300 genes most positively correlated with FBXO45 were screened into the enrichment analysis. The functional enrichment results showed that FBXO45 was mainly associated with proteasomal protein catabolic process and the regulation of DNA (deoxyribonucleic acid) metabolic processing in the biological process (BP) category (Figure 5C); spindle, chromosomal region, and focal adhesion in the cellular component (CC) category (Figure 5D); and ATPase (adenosine triphosphate) activity and ubiquitin-protein transferase activity terms in the molecular function (MF) category (Figure 5E). KEGG analysis showed that FBXO45 might participate in the cell cycle, as well as the Wnt (Wingless/Integrated), mTOR (mammalian target of rapamycin), and Hippo signaling pathways (Figure 5F).
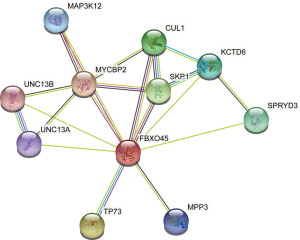
PPI network construction
We further constructed the PPI network using the STRING database. A total of 10 proteins were closely associated with FBXO45 protein, of which MYCBP2 and SKP1 were most associated with FBXO45 (Figure 6).
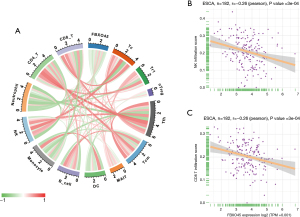
Immune cell infiltration analysis
To verify the immune-regulation function of FBXO45, we downloaded the infiltration levels of 24 immune cells from the ImmuCellAI database, and compared the immune cell infiltration levels in the high and low FBXO45 expression groups. We found that the infiltration levels of CD4 T cells, CD8 T cells, and NK cells were lower in the high FBXO45 expression group in ESCA (Figure 7).

We also performed correlation analysis between FBXO45 expression and the immune cell infiltration level (Figure 8A). Through the correlation analysis, we observed that FBXO45 expression was negatively correlated with immune killer cells, including NK cells and CD8+ T cells, in ESCA (Figure 8B,8C). These results revealed that patients with high FBXO45 expression might be in a relatively immunosuppressive environment. FBXO45 was mainly expressed in the cytoplasm of ESCA cells (Figure 9).

Discussion
ESCA is a common gastrointestinal tumor. Although some progress has been made in cancer-related treatment technology in recent years, the 5-year survival rate of ESCA is still very low (11-13). Recent studies have shown that the remodeling of the TIME by cancer cells plays an important role in the development of ESCA, thereby weakening the response of ESCA patients to treatment and leading to worse survival status (14,15). Thus, identification of essential genes that could affect the TIME is urgently needed.
FBXO45, a substrate-recognition subunit of E3 ligases, is a member of the F-box protein family. FBXO45 is reportedly involved in the progression of numerous tumors. However, the role of FBXO45 in ESCA and its correlation with the TIME remains unclear. In our study, we first assessed the expression of FBXO45 and found that FBXO45 expression was higher in tumor tissues compared with normal tissues in ESCA. In addition, FBXO45 expression was relatively higher in worse tumor stages. We also observed that FBXO45 was highly expressed in esophageal squamous cell carcinoma, compared to esophageal adenocarcinoma.
Furthermore, we conducted enrichment analysis of FBXO45. The functional enrichment results revealed that FBXO45 was mainly associated with proteasomal protein catabolic process and regulation of DNA metabolic processing in BP category; spindle, chromosomal region, and focal adhesion in the CC category; and ATPase activity and ubiquitin-protein transferase activity terms in the MF category. KEGG analysis revealed that FBXO45 might participate in the cell cycle, as well as the Wnt, mTOR, and Hippo signaling pathways. These results suggested that FBXO45 might be an oncogene that is involved in multiple cancer-promoting pathways.
Through the PPI network, we found that SKP1 was closely associated with the FBXO45 protein. It was previously reported that SKP1 is an oncogene in many tumor types, including colorectal (16), breast (17), skin (18), lung (19), and bladder (20) cancers. SKP1 is also involved in ESCA (21-23). We believe that the interaction between FBXO45 and SKP1 may be one of the mechanisms through which FBXO45 promotes the progression of ESCA.
In addition, we further explored the role of FBXO45 in the TIME. Our results suggested that FBXO45 expression was negatively correlated with immune killer cells, including NK cells and CD8+ T cells, in ESCA. These results demonstrated that patients with high FBXO45 expression might be in relatively immunosuppressive environment.
In conclusion, our findings showed that FBXO45 acts as an oncogene in ESCA. High FBXO45 expression may contribute to an immunosuppressive microenvironment in ESCA.
Acknowledgments
First and foremost, Jian Zhang shall extend his thanks to the funding of project of Zhejiang Provincial Key Laboratory of Microinvasive Diagnosis, Treatment and Rapid Rehabilitation of Digestive System Tumor. Last but not least, Jian Zhang would like to thank all his friends, especially his wife, for their encouragement and support.
Funding: This study was supported by the project of Zhejiang Provincial Key Laboratory of Microinvasive Diagnosis, Treatment and Rapid Rehabilitation of Digestive System Tumor.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-662
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-662). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Short MW, Burgers KG, Fry VT. Esophageal Cancer. Am Fam Physician 2017;95:22-8. [PubMed]
- Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol 2015;21:7933-43. [Crossref] [PubMed]
- Bollschweiler E, Plum P, Mönig SP, et al. Current and future treatment options for esophageal cancer in the elderly. Expert Opin Pharmacother 2017;18:1001-10. [Crossref] [PubMed]
- Jiang P, Li Y, Xu Z, et al. A signature of 17 immune-related gene pairs predicts prognosis and immune status in HNSCC patients. Transl Oncol 2021;14:100924. [Crossref] [PubMed]
- Altorki NK, Markowitz GJ, Gao D, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer 2019;19:9-31. [Crossref] [PubMed]
- Rojas A, Araya P, Gonzalez I, Morales E. Gastric Tumor Microenvironment. Adv Exp Med Biol 2020;1226:23-35. [Crossref] [PubMed]
- Höpken UE, Rehm A. Targeting the Tumor Microenvironment of Leukemia and Lymphoma. Trends Cancer 2019;5:351-64. [Crossref] [PubMed]
- Wang K, Qu X, Liu S, et al. Identification of aberrantly expressed F-box proteins in squamous-cell lung carcinoma. J Cancer Res Clin Oncol 2018;144:1509-21. [Crossref] [PubMed]
- Kogure N, Yokobori T, Ogata K, et al. Low Expression of FBXO45 Is Associated with Gastric Cancer Progression and Poor Prognosis. Anticancer Res 2017;37:191-6. [Crossref] [PubMed]
- Dahlem C, Barghash A, Puchas P, et al. The Insulin-Like Growth Factor 2 mRNA Binding Protein IMP2/IGF2BP2 is Overexpressed and Correlates with Poor Survival in Pancreatic Cancer. Int J Mol Sci 2019;20:3204. [Crossref] [PubMed]
- Zhou N, Hofstetter WL. Prognostic and therapeutic molecular markers in the clinical management of esophageal cancer. Expert Rev Mol Diagn 2020;20:401-11. [Crossref] [PubMed]
- Anandavadivelan P, Lagergren P. Cachexia in patients with oesophageal cancer. Nat Rev Clin Oncol 2016;13:185-98. [Crossref] [PubMed]
- Xu HT, Miao J, Liu JW, et al. Prognostic value of circulating tumor cells in esophageal cancer. World J Gastroenterol 2017;23:1310-8. [Crossref] [PubMed]
- Baba Y, Nomoto D, Okadome K, et al. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci 2020;111:3132-41. [Crossref] [PubMed]
- Huang TX, Fu L. The immune landscape of esophageal cancer. Cancer Commun (Lond) 2019;39:79. [Crossref] [PubMed]
- Tian C, Lang T, Qiu J, et al. SKP1 promotes YAP-mediated colorectal cancer stemness via suppressing RASSF1. Cancer Cell Int 2020;20:579. [Crossref] [PubMed]
- Tian Z, He W, Tang J, et al. Identification of Important Modules and Biomarkers in Breast Cancer Based on WGCNA. Onco Targets Ther 2020;13:6805-17. [Crossref] [PubMed]
- Xie CM, Wei W, Sun Y. Role of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases in skin cancer. J Genet Genomics 2013;40:97-106. [Crossref] [PubMed]
- Liu YQ, Wang XL, Cheng X, et al. Skp1 in lung cancer: clinical significance and therapeutic efficacy of its small molecule inhibitors. Oncotarget 2015;6:34953-67. [Crossref] [PubMed]
- Wu S, Yang J, Xu H, et al. Circular RNA circGLIS3 promotes bladder cancer proliferation via the miR-1273f/SKP1/Cyclin D1 axis. Cell Biol Toxicol 2021; Epub ahead of print. [Crossref] [PubMed]
- Dong S, Zhao J, Wei J, et al. F-box protein complex FBXL19 regulates TGFβ1-induced E-cadherin down-regulation by mediating Rac3 ubiquitination and degradation. Mol Cancer 2014;13:76. [Crossref] [PubMed]
- Lian Z, Lee EK, Bass AJ, et al. FBXO4 loss facilitates carcinogen induced papilloma development in mice. Cancer Biol Ther 2015;16:750-5. [Crossref] [PubMed]
- Liu J, Lv L, Gong J, et al. Overexpression of F-box only protein 31 predicts poor prognosis and deregulates p38α- and JNK-mediated apoptosis in esophageal squamous cell carcinoma. Int J Cancer 2018;142:145-55. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


