
Comprehensive analysis of prognostic value and immune infiltration of calpains in pancreatic cancer
Introduction
Calpains (CAPNs) are intracellular calcium-activated neutral cysteine proteinases involved in multiple cellular processes (1-3). They are also known to mediate cancer initiation, progression, and metastasis by catalyzing the proteolysis of numerous substrates (4,5). In total, 16 members of the human CAPN family have been identified by the Human Genome Project and categorized into classical (CAPN1–3, 8, 9, and 11–14) and non-classical (CAPN5–7, 10, 15, and 16) based on the primary sequence and ubiquitous (CAPN1, 2, 5, 7, 10, 13–15, and S1) or tissue-specific (CAPN3, 6, 8, 9, 11, and 12) localizations (6). Previous studies suggested that aberrant expression of CAPNs is related to many cancer types. For instance, increased expression of CAPN1 to renal carcinomas (7), of CAPN2 to colorectal cancer and prostate cancer (8,9), and of CAPN6 to uterine sarcomas, carcinosarcomas, and uterine cervical neoplasia (10,11). On the other hand, decreased expression of CAPN9 is associated with gastric cancer development (12). CAPNs can modulate key molecules in signaling pathways, including the protein kinase Ciota, focal adhesion kinase (FAK), tumor suppressor protein p53, hypoxia-inducible factor 1, and epidermal growth factor receptor, contributing to tumor formation, development, progression, proliferation, differentiation, invasion, and metastasis (13-16).
Pancreatic cancer (PC) is one of the most common malignancies worldwide, and only 20% of the cases are resectable due to the lack of obvious clinical symptoms in the early phase (17,18). Even after curative surgery, the recurrence rates are high, while long-term survival rates are shorter than other cancers (19). Aggressive biological behavior characterizes PC because of its high extracellular stromal components that generate a hypovascular and hypoxic microenvironment. It is also characterized by poor prognosis, because the underlying mechanisms of carcinogenesis and immune response remain unclear (20).
Earlier studies have shown that some CAPNs are closely related to PC (21-23). For instance, a study by Yu et al. (22) showed that CAPN1 expression was higher in tumor tissue than that in normal tissue, and overexpressed CAPN1 was associated with advanced tumor stage and tumor metastasis. Otherwise, this study also showed that CAPN1 overexpression was correlated with poor prognosis, which can promote tumor cell proliferation, invasion, and migration in in vitro study. However, the mechanism of how CAPN1 functions was not elucidated clearly. Other studies also showed that part of CAPNs played important roles in PC, which indicated that calpains family involved in the biological process of PC, but the distinct functions have not been elucidated either. Considering the special tumor microenvironment in PC, there has been increasingly more attention to the tumor-infiltrating immune cells. As far as we know, there is no study investigating the relationship between CAPNs and immune infiltration before. Thanks to the development of bioinformatic tools, we can predict the possible mechanism of immune infiltration in PC according to the different expression levels of CAPNs. Here, we aimed to combine previously published gene expression or copy number variation data to determine the expression profiles, potential functions, distinct prognostic values, and immune infiltration status of CAPNs in PC. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-705).
Methods
This study was carried out with adherence to the stipulations of the Declaration of Helsinki (as revised in 2013). Both ethical approval and patient consent were not required, because this study did not include any experiments involving animal or human subjects.
Oncomine
The transcription levels of CAPNs in different cancers were determined through analysis in Oncomine (https://www.oncomine.org) (24). In this study, the student’s t-test was used to compare the difference in the expression of CAPNs in PC. A P value <0.01, a fold change of 2, and a gene rank in the top 10% were set as the significance thresholds.
GEPIA database
GEPIA (http://gepia.cancer-pku.cn) is an interactive web server using a standard processing pipeline, and consists of thousands of tumors and normal tissue samples data from the Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) projects (25). In this study, differential gene expression analysis was performed to compare tumor/normal tissues and pathological stage analysis through GEPIA. The Student’s t-test was used to generate a P value for the expression or pathological stage analysis. A statistically significant difference was set as a P value <0.05.
Kaplan-Meier plotter
Kaplan-Meier plotter (http://kmplot.com/analysis) is an online database that contains information about the association of 54,000 gene expression with the survival of patients with 21 different cancers (26). This study analyzed the prognostic value of the mRNA expression of CAPNs in PC patients using this tool. Information regarding the number-at-risk cases, median mRNA expression levels, HRs, 95% CIs and P values was presented. A statistically significant difference was set as a P value <0.05.
The Human Protein Atlas
The Human Protein Atlas (https://www.proteinatlas.org/) is an online database that contains transcriptome profiling data and immunohistochemistry images for over 8,000 patients and 17 types of cancers (27). In this study, we checked the immunohistochemical images to directly compare the protein expression of different CAPNs in human normal pancreatic tissue and PC tissues, and analyze the CAPNs protein expression patterns.
TCGA and cBioPortal
TCGA (https://www.cancer.gov/tcga), a landmark cancer genome project, includes sequencing and pathological data on 33 different cancers (28). The cBioPortal (https://www.cbioportal.org) database contains all data in TCGA and is continuously updated (29). In this study, the PC datasets, including data from 850 cases with pathology reports, were selected for further analyses of CAPNs using cBioPortal. The CAPNs gene alteration in PC was obtained, including genetic mutations, gene fusions, gene amplifications, deep deletions, and multiple alterations.
STRING and GeneMANIA
STRING (https://string-db.org) is a website about predicting protein-protein interactions (PPI) networks based on direct (physical) and indirect (functional) associations (30). Cytoscape is a popular open-source software tool that can visualize biomedical networks consisting of proteins, genes, and other types of interactions. In this study, a PPI network analysis was conducted to analyze the differential expression of CAPNs and potential interactions through STRING and Cytoscape software. GeneMANIA (http://www.genemania.org) is a resource-rich website containing gene information, predicting a functional interaction network to explore the association between genes and data sets (31). In this study, we used it to the weight that indicates the predictive value of CAPNs.
GO and KEGG
GO and KEGG analyses are used to analyze the functional enrichment of discrepant CAPNs in PC and their high-related co-expressed molecular, which are performed in the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov) (32,33). In details, GO analyses contain biological process, cellular component, and molecular function.
Single sample gene set enrichment analysis (ssGSEA)
The ssGSEA method quantified the tumor-infiltrating immune cells using the GSVA package in R software (34,35). The ssGSEA applies gene signatures expressed by immune cell populations to individual cancer samples. The deconvolution approach used in our study including 24 immune cells that are involved in immunity, including activated DC (aDC), B cells, CD8 T cells, Cytotoxic cells, DC, Eosinophils, immature DC (iDC), Macrophages, Mast cells, Neutrophils, NK CD56bright cells, NK CD56dim cells, NK cells, Plasmacytoid DC (pDC), T cells, T helper (Th) cells, T central memory (Tcm), T effector memory (Tem), T follicular helper (Tfh), T gamma delta (Tgd), Th1 cells, Th17 cells, Th2 cells, and regulatory T (Treg) cells. In this study, we conducted the ssGSEA algorithm to explore the relationship between the CAPNs and the infiltration of immune cells based on the dataset of PC (TCGA, Firehose Legacy). The correlation between CAPNs and tumor-infiltrating immune cells was assessed by the Spearman test.
The statistical analysis
Oncomine and GEPIA databases provided t-test analysis. Kaplan-Meier plotter online tool was used for statistical analyses of hazard ratios and survival rates. A log-rank test was performed to evaluate the equality of survival curves between different patient groups.
Results
Transcriptional levels of CAPNs
Using the Oncomine database, we identified 16 members of CAPNs related to PC and compared their transcriptional levels in cancer and normal tissues (Figure 1; Table 1). Compared to normal tissue, CAPN1 was significantly upregulated in cancer tissues using the Logsdon’s (2.074-fold change) and Pei’s (2.050-fold change) datasets. CAPN9 was found to be downregulated in cancer tissue according to Lacobuzio-Donahue’s dataset (12.176-fold change) and Pei’s dataset (2.900-fold change). CAPN2, CAPNS1, and CAPN5 were also highly expressed compared to normal tissue, according to Badea’s dataset (2.027-fold change), Logsdon’s dataset (2.230-fold change), and Pei’s dataset (2.222-fold change). CAPN8 and CAPN12 were found overexpressed in Pei’s dataset (12.607- and 2.189-fold change, respectively).
Table 1
| Gene | Fold change | P value | t test | Source |
|---|---|---|---|---|
| CAPN1 | 2.074 | 2.09E-6 | 7.713 | Logsdon pancreas |
| 2.050 | 1.63E-7 | 5.956 | Pei pancreas | |
| CAPN2 | 2.027 | 9.17E-11 | 7.721 | Badea pancreas |
| CAPNS1 (CAPN4) | 2.230 | 1.71E-6 | 7.808 | Logsdon pancreas |
| CAPN5 | 2.562 | 1.10E-7 | 6.120 | Pei pancreas |
| CAPN8 | 12.607 | 5.71E-11 | 8.474 | Pei pancreas |
| CAPN9 | 12.176 | 7.13E-5 | 5.305 | Lacobuzio-Donahue pancreas 2 |
| 2.900 | 1.43E-7 | 5.925 | Pei pancreas | |
| CAPN12 | 2.189 | 2.15E-6 | 5.176 | Pei pancreas |
Relationship between CAPN expression and clinicopathological parameters of PC
Using the GEPIA database, we compared the transcriptional levels of CAPNs in cancer and normal tissues. The results indicated that the expression levels of CAPN1, 2, 5, 6, 8–10, 12, and S1 were higher in cancer tissues than in normal ones (Figure 2). We found that CAPN1, 5, 8, and 12 overexpression was correlated with advanced-stage tumors (Figure 3).
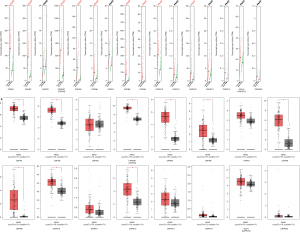
Prognostic value of CAPN expression
Using the Kaplan-Meier plotter, we analyzed the correlations between the transcriptional expression of CAPNs and clinical outcomes of PC patients. The overall survival (OS) and recurrence-free survival (RFS) curves are shown in Figure 4. The transcriptional levels of CAPN1–3, 5, and 8 were negatively correlated with OS, and those of CAPN1, 2, 5, and 8 were negatively correlated with RFS. In contrast, the expression of CAPN10 was positively correlated with OS and RFS. Thus, CAPN1, 2, 5, 8, and 10 could be used as predictive markers for clinical outcomes in patients with PC (Figure 5). In subgroup analysis, we investigated the correlation between the transcriptional expression of CAPN1, 2, 5, 8, and 10 with clinicopathological parameters (i.e., sex, age, smoking, diabetes, and clinical stage). The results showed that CAPN1, 2, and 5 were good prognostic biomarkers in males and females. CAPN8 was only found in female patients, and CAPN10 was only present in males (Table 2). All five CAPNs were significantly correlated with prognosis in young patients, while CAPN1 was also a good prognostic biomarker for elderly patients (Table 3). All five CAPNs were correlated with PC prognosis in non-smokers, and CAPN10 was also a prognostic biomarker for smokers (Table 4). Similarly, all five CAPNs were prognostic biomarkers for patients without a history of diabetes, and CAPN10 was also correlated with PC prognosis in patients with a history of diabetes (Table 5). Due to the limited number of patients with stage III or IV PC, we only analyzed the predictive effects of CAPNs in patients with stage I and II PC. The results showed that CAPN1 and 5 were the only prognostic biomarkers in patients with stage II PC, while CAPN2, 8, and 10 were correlated with stages I and II PC (Table 6).

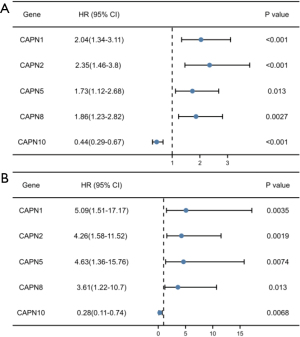
Table 2
| Gene | Sex | Cases | HR (95% CI) | P value |
|---|---|---|---|---|
| CAPN1 | Female | 80 | 2.39 (1.14–4.98) | 0.017 |
| Male | 97 | 2.14 (1.17–3.89) | 0.011 | |
| CAPN2 | Female | 80 | 2.18 (1.16–4.10) | 0.013 |
| Male | 97 | 3.01 (1.40–6.47) | 0.0031 | |
| CAPN5 | Female | 80 | 1.95 (1.06–3.59) | 0.028 |
| Male | 97 | 1.99 (1.11–3.58) | 0.019 | |
| CAPN8 | Female | 80 | 2.61 (1.41–4.83) | 0.0015 |
| Male | 97 | 1.72 (0.96–3.10) | 0.066 | |
| CAPN10 | Female | 80 | 0.57 (0.30–1.06) | 0.072 |
| Male | 97 | 0.32 (0.17–0.57) | 6.5e-05 |
Table 3
| Gene | Age | Cases | HR (95% CI) | P value |
|---|---|---|---|---|
| CAPN1 | ≤65 | 92 | 3.27 (1.36−7.84) | 0.008 |
| >65 | 85 | 2.16 (1.04−4.49) | 0.038 | |
| CAPN2 | ≤65 | 92 | 3.66 (1.62−8.26) | 0.002 |
| >65 | 85 | 1.92 (0.89−4.12) | 0.096 | |
| CAPN5 | ≤65 | 92 | 2.99 (1.25−7.18) | 0.014 |
| >65 | 85 | 1.35 (0.73−2.50) | 0.339 | |
| CAPN8 | ≤65 | 92 | 2.21 (1.21−4.01) | 0.009 |
| >65 | 85 | 1.08 (0.61−1.94) | 0.783 | |
| CAPN10 | ≤65 | 92 | 0.33 (0.18−0.62) | 0.001 |
| >65 | 85 | 0.61 (0.34−1.09) | 0.097 |
Table 4
| Gene | Smoker | Cases | HR (95% CI) | P value |
|---|---|---|---|---|
| CAPN1 | Yes | 79 | 1.40 (0.75−2.61) | 0.285 |
| No | 65 | 2.17 (1.05−4.48) | 0.037 | |
| CAPN2 | Yes | 79 | 0.56 (0.26−1.21) | 0.139 |
| No | 65 | 6.12 (2.13−17.57) | 0.001 | |
| CAPN5 | Yes | 79 | 1.51 (0.82−2.80) | 0.186 |
| No | 65 | 2.59 (1.20−5.57) | 0.015 | |
| CAPN8 | Yes | 79 | 1.45 (0.79−2.64) | 0.226 |
| No | 65 | 3.21 (1.47−7.03) | 0.003 | |
| CAPN10 | Yes | 79 | 0.36 (0.20−0.67) | 0.001 |
| No | 65 | 0.45 (0.22−0.94) | 0.034 |
Table 5
| Gene | History of diabetes | Cases | HR (95% CI) | P value |
|---|---|---|---|---|
| CAPN1 | Yes | 38 | 9.28 (1.20−71.55) | 0.032 |
| No | 108 | 1.84 (1.09−3.11) | 0.023 | |
| CAPN2 | Yes | 38 | 3.69 (1.17−11.60) | 0.025 |
| No | 108 | 2.32 (1.17−4.60) | 0.016 | |
| CAPN5 | Yes | 38 | 0.90 (0.33−2.45) | 0.835 |
| No | 108 | 2.23 (1.28−3.91) | 0.005 | |
| CAPN8 | Yes | 38 | 0.60 (0.20−1.80) | 0.366 |
| No | 108 | 2.55 (1.49−4.37) | 0.001 | |
| CAPN10 | Yes | 38 | 0.18 (0.05−0.63) | 0.007 |
| No | 108 | 0.49 (0.29−0.82) | 0.007 |
Table 6
| Gene | Stage | Cases | HR (95% CI) | P value |
|---|---|---|---|---|
| CAPN1 | 1 | 21 | >10.00 (0–lnf) | 0.051 |
| 2 | 146 | 1.67 (1.07–2.61) | 0.022 | |
| 3 | 3 | NA | NA | |
| 4 | 5 | NA | NA | |
| CAPN2 | 1 | 21 | 5.83 (1.12–30.27) | 0.018 |
| 2 | 146 | 1.72 (1.03–2.86) | 0.034 | |
| 3 | 3 | NA | NA | |
| 4 | 5 | NA | NA | |
| CAPN5 | 1 | 21 | >10.00 (0–lnf) | 0.023 |
| 2 | 146 | 0.66 (0.41–1.06) | 0.086 | |
| 3 | 3 | NA | NA | |
| 4 | 5 | NA | NA | |
| CAPN8 | 1 | 21 | 10.36 (1.22–87.91) | 0.0088 |
| 2 | 146 | 0.59 (0.36–0.96) | 0.033 | |
| 3 | 3 | NA | NA | |
| 4 | 5 | NA | NA | |
| CAPN10 | 1 | 21 | 0.09 (0.01–0.73) | 0.0047 |
| 2 | 146 | 0.53 (0.35–0.83) | 0.0042 | |
| 3 | 3 | NA | NA | |
| 4 | 5 | NA | NA |
Protein expression of CAPNs
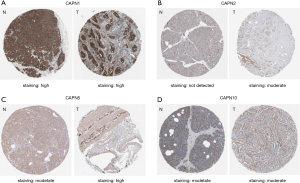
We checked the immunohistochemical images of CAPNs with a predictive value in the Human Protein Atlas (Figure 6). In tumor tissue, the staining of CAPN1 and CAPN5 was high, whereas that of CAPN2 and CAPN10 was moderate. In normal tissue, the staining of CAPN1 was high, CAPN5 and CAPN10 were moderate, whereas CAPN2 was not detected. Immunohistochemical images of CAPN8 were unavailable.
Gene mutations and interaction analyses of CAPNs
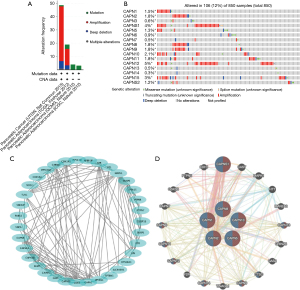
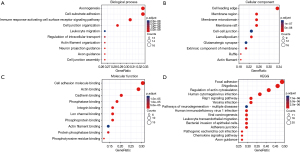
Using the cBioPortal database, we analyzed the genetic alterations of CAPNs in patients with PC. Overall, mRNA amplification, mutation, and deep deletion were the most important factors affecting alterations in PC (Figure 7A). CAPNs were altered in 850 tissue samples of 849 patients with PC, accounting for 12% (Figure 7B). The genetic alterations in CAPNs for PC varied from 0.1% to 5% (CAPN1, 1.5%; CAPN2, 1.8%; CAPN3, 0.6%; CAPNS1, 4%; CAPN5, 1.3%; CAPN6, 0.9%; CAPN7, 0.5%; CAPN8, 1.8%; CAPN9, 1.8%; CAPN10, 2.1%; CAPN11, 1.8%; CAPN12, 5%; CAPN13, 0.5%; CAPN14, 0.3%; CAPN15, 3%; CAPNS2, 1.2%). A protein-protein interaction (PPI) network analysis for CAPN1, 2, 5, 8, and 10 resulted in 35 nodes and 140 edges (Figure 7C). Using the GeneMANIA database, we found that five central nodes surrounded 20 nodes, revealing genes related to physical interaction, co-expression, prediction, co-localization, genetic interactions, pathways, and shared protein domains (Figure 7D). Furthermore, the corresponding biological functions of the five differentially expressed CAPNs, and their highly related co-expressed genes were analyzed using the DAVID database. The results of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched signaling pathways analyses are shown in Figure 8.
Tumor-infiltrating immune cell of CAPNs
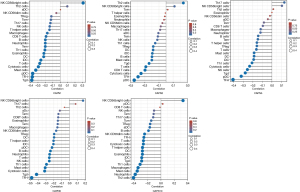
Tumor-infiltrating immune cells are involved in the proliferation and progression of cancer cells. We used the ssGSEA method to explore the correlation between CAPN expression and immune cell infiltration (Figure 9). The expression of CAPN1 was correlated with the infiltration of B cells, CD8 T cells, cytotoxic cells, DCs, eosinophils, iDCs, macrophages, mast cells, neutrophils, CD56 bright NK cells, pDCs, T cells, TEM, Tfh cells, Tgd cells, and Th1 cells. The expression of CAPN2 was positively correlated with the infiltration of pDCs, Tfh, cytotoxic cells, CD8 T cells, T cells, Th2 cells, Tgd cells, mast cells, B cells, iDCs, DCs, Treg cells, and Th1 cells. The expression of CAPN5 was positively correlated with the infiltration of Tfh, pDCs, Tgd cells, NK cells, cytotoxic cells, Th1 cells, iDCs, DCs, mast cells, T cells, Treg cells, and Th cells. The expression of CAPN8 was correlated with the infiltration of almost all immune cells, and that of CD8 T cells, Tcm cells, aDCs, Th17 cells, and Th2 cells. The expression of CAPN10 was correlated with the infiltration of all immune cells, except for Tcm, Th17 cells, Tem, NK cells, pDCs, and CD8 T cells.
Discussion
As a widespread post-translational modification of proteins, the CAPN-mediated cleavage regulates numerous cellular processes, including proliferation, differentiation, cytoskeletal reorganization, and apoptosis. The Human Genome Project has discovered 16 members of the CAPN family. Based on the molecular structure, CAPN1–3 and CAPN5–16 are the large catalytic subunits that contain domains DI, DII, DIII, and DVI, whereas CAPNS1 and CAPNS2 are the small regulatory subunits that contain domains DV and DVI. CAPNs are classified into classical (CAPN1–3, 8, 9, 11–14) and non-classical (CAPN5–7, 10, 15, 16), based on the presence or absence of domain DVI, respectively. CAPNs modulate key molecules in several signaling pathways that contribute to carcinogenesis or malignant metastasis, with a potential prognostic value in various malignancies. However, their role and prognostic value in PC remain unclear. This is the first report on the bioinformatic analysis of CAPNs to explore their prognostic value in PC.
Our results showed that the expression of CAPNs significantly increased or decreased in PC, indicating a potential prognostic value. CAPN1, 2, 5, and 8 were prognostic biomarkers negatively correlated with OS and RFS, while CAPN 10 was positively correlated with the same parameters. Moreover, the genetic alternation rate of CAPNs in patients with PC was estimated at 12.0%.
CAPN1 and CAPN2 were first discovered in 1964 and share high sequence similarity of 62% in humans. Previous studies showed that CAPN1 is overexpressed in various types of malignancies. For instance, CAPN1 and CAPN2 are overexpressed in colorectal and prostate cancers (36,37). Both CAPN1 and CAPN2 are expressed in small cell lung cancer and non-small cell lung cancer (NSCLC), and their upregulation increases the migration and invasion of lung cancer cells. Chen et al. demonstrated that CAPN1 promotes malignant behavior in lung adenocarcinoma (22,38). A previous study described that CAPN2 up-regulates the FAK-phosphatidylinositol 3-kinase /protein kinase B and extracellular signal-regulated kinase 1/2 signaling pathways to facilitate lung cancer metastasis (39). Trillsch et al. demonstrated that CAPN1 cleaves the key epithelial-to-mesenchymal transition marker, E-cadherin, and up-regulates the canonical Wnt/β-catenin and non-canonical Wnt5A signaling pathways to facilitate tumor metastasis in advanced epithelial ovarian cancer (40). Based on our findings, the transcriptional expression of CAPN1 and CAPN2 was significantly upregulated in PC tissues compared with that in normal tissues. CAPN1, but not CAPN2, was positively correlated with the advanced tumor stage, and both were negatively correlated with OS and RFS; thus, CAPN1 and CAPN2 have a potential prognostic value in PC.
CAPN3 is predominantly expressed in skeletal muscle, and the protein structure differs from conventional CAPNs in three sequences (1). Previous studies showed that CAPN3 variants play a pro-apoptotic role in melanoma cells, and their downregulation contributes to cancer progression (41,42). Our results showed that the transcriptional expression of CAPN3 was not significantly upregulated in PC tissues compared with that in normal ones. Thus, CAPN3 might not be related to PC progression.
CAPNS1 mediates cell death through different cellular signaling pathways. It is known to prevent apoptosis via the PI3K/Akt pathway (43) and promote apoptosis via the calpain-protein phosphatase 2A (PP2A)-Akt-forkhead box O (FoxO) pathway (44). Previous studies have demonstrated that CAPNS1 promotes tumor progression in NSCLC (45), gastric cancer (46), nasopharyngeal carcinoma (47), and intrahepatic cholangiocarcinoma (48). Our results showed that the transcriptional expression of CAPNS1 was significantly higher in PC tissues than in normal tissues, but no correlation was found with OS or RFS.
Of the non-classical CAPNs that lack domain DVI, CAPN5 is highly expressed in the central nervous system, retina, and gastrointestinal tract, but its role in various cancers remains unclear (49-51). CAPN6 is highly expressed in the placenta and embryos, correlated with malignant diseases, such as leiomyosarcoma, endometrial stromal sarcoma, and cervical cancer, and promotes angiogenesis by increasing the secretion of vascular endothelial growth factor (10,49,52-55). CAPN7 promotes the migration and invasion of human endometrial stromal cells, but its role in cancer has not been fully elucidated (56). CAPN10 plays a role in diabetes mellitus and is recognized as a potential prognostic biomarker for esophageal squamous cell carcinoma (57-59). CAPN15 plays a critical role in the development of eye disorders (60). Our study revealed that the transcriptional expression of CAPN5, 6, and 10 was significantly higher in PC tissues than in normal tissues. Besides, the expression of CAPN5 was negatively correlated with OS and RFS, while the expression of CAPN10 was positively correlated with the same parameters. Therefore, CAPN5 and CAPN10 could be used as prognostic biomarkers for PC, and the protective mechanism of the latter requires further investigation.
CAPN8 and 9 that show highly conserved gene organization of other typical CAPN large subunits are predominantly expressed in the stomach and the gastrointestinal tract (61,62). It is known that CAPN8 is correlated with the progression of lung squamous cell carcinoma, while CAPN9 is considered a prognostic biomarker for clinical outcomes of gastric cancer (63). Our study revealed that CAPN8 and CAPN9 were negatively correlated with OS and RFS, and thus could be used as prognostic biomarkers for survival in PC.
CAPN11 has a high sequence similarity to CAPN1 and CAPN2 and is overexpressed in the testis (64,65); however, limited information on its role in cancers. CAPN12 is predominantly expressed in skin tissues (66,67), and little is known about its function in various malignant diseases. We found that CAPN12 had significantly higher expression in PC tissues than in normal ones. Thus, further studies are needed to investigate the underlying molecular mechanism of CAPN12 in PC.
CAPN13 is considered the parent of all classical CAPNs, and selectively, lowly expressed in the testis and lungs (68,69). CAPN14 has a sequence similarity of 36% with CAPN13 and is not detected in any of the previously studied tissues. However, it has been reported to be correlated with eosinophilic esophagitis (68,70,71). In our study, CAPN13 was not differentially expressed in PC tissues, while CAPN14 was undetectable.
PC is characterized by extensive desmoplasia, hypoperfusion, and an immunosuppressive microenvironment. The tumor microenvironment includes cells associated with innate and adaptive immunity (72). Host immune responses to tumor cells are represented by tumor-infiltrating immune cells (73,74). A previous study reported that tumor-infiltrating Tregs are associated with poor prognosis in patients with PC (75). Besides, tumor-infiltrating CD4+ T and CD8+ T cells are positively correlated with survival (76). De Monte et al. (77) found that Th2 rather than Th1 cells predominantly infiltrated in PC, and that their ratio is an independent prognostic biomarker. Therefore, the systematic analysis of tumor-infiltrating immune cells is important for indicating the effects of specific cell combinations in the immune microenvironment. Here, we found that CAPN1, 2, 5, and 8 were highly correlated with tumor-infiltrating Tfh cells, which are a specialized subset of CD4+ T cells, and play a crucial role in regulating antigen-specific B cell responses. In breast cancer, the differentiation of tumor-infiltrating CXCL13-producing Tfh cells plays a role in converting Treg-mediated immune suppression to de novo activation of adaptive anti-tumor humoral responses, and Tfh cell activation in B cells induces anti-tumor response (78,79). We found that CAPN10 is correlated with the tumor-infiltrating Th2 cells and thus could be used as a biomarker for response to immune checkpoint inhibitors.
Enrichment analysis indicated that the functions of CAPNs and the related differential proteins are correlated with axonogenesis, cell-substrate adhesion, immune response-activating cell surface receptor signaling pathway, and cell junction organization in PC, results consistent with those reported previously (4). Furthermore, KEGG enrichment analysis that prognostic CAPNs in PC were associated with focal adhesion, regulation of actin cytoskeleton, and the rap1 signaling pathway suggests their role in modulating tumor progression.
Our study had some limitations: (I) the number of enrolled patients in the used databases was relatively small, and (II) all the data were analyzed based on online databases. Therefore, extensive basic and clinical research is needed to validate our findings.
Conclusions
Here, we performed a comprehensive bioinformatic analysis to investigate the expression levels and prognostic value of CAPNs in PC. Two main subgroups were identified; the first (CAPN1, 2, 5, 8, and 12) was negatively correlated with OS and RFS and correlated with tumor-infiltrating Tfh cells, while the second (CAPN10) was positively correlated with OS and RFS, as well as with tumor-infiltrating Th2 cells. Thus, some members of the CAPN family could be used as prognostic biomarkers and help improve individualized treatment strategies in patients with PC.
Acknowledgments
We would like to acknowledge the Oncomine, GEPIA, Kaplan-Meier Plotter, THPA, TCGA, cBioPortal, STRING, GeneMANIA, and DAVID databases used free of charge. Meanwhile, we would like to thank Editage (www.editage.cn) for English language editing.
Funding: The present study was supported by the Project of Sichuan Provincial Science and Technology Department (2018JY0489) and the Project of Sichuan Provincial Health Commission (20PJ150).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-705
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jgo-21-705). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was carried out with adherence to the stipulations of the Declaration of Helsinki (as revised in 2013). Both ethical approval and patient consent were not required, because this study did not include any experiments involving animal or human subjects.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sorimachi H, Ishiura S, Suzuki K. Structure and physiological function of calpains. Biochem J 1997;328:721-32. [Crossref] [PubMed]
- Glading A, Lauffenburger DA, Wells A. Cutting to the chase: calpain proteases in cell motility. Trends Cell Biol 2002;12:46-54. [Crossref] [PubMed]
- Jánossy J, Ubezio P, Apáti A, et al. Calpain as a multi-site regulator of cell cycle. Biochem Pharmacol 2004;67:1513-21. [Crossref] [PubMed]
- Nian H, Ma B. Calpain-calpastatin system and cancer progression. Biol Rev Camb Philos Soc 2021;96:961-75. [Crossref] [PubMed]
- Moretti D, Del Bello B, Allavena G, et al. Calpains and cancer: friends or enemies? Arch Biochem Biophys 2014;564:26-36. [Crossref] [PubMed]
- Storr SJ, Carragher NO, Frame MC, et al. The calpain system and cancer. Nat Rev Cancer 2011;11:364-74. [Crossref] [PubMed]
- Braun C, Engel M, Seifert M, et al. Expression of calpain I messenger RNA in human renal cell carcinoma: correlation with lymph node metastasis and histological type. Int J Cancer 1999;84:6-9. [Crossref] [PubMed]
- Lakshmikuttyamma A, Selvakumar P, Kanthan R, et al. Overexpression of m-calpain in human colorectal adenocarcinomas. Cancer Epidemiol Biomarkers Prev 2004;13:1604-9. [PubMed]
- Mamoune A, Luo JH, Lauffenburger DA, et al. Calpain-2 as a target for limiting prostate cancer invasion. Cancer Res 2003;63:4632-40. [PubMed]
- Lee SJ, Choi YL, Lee EJ, et al. Increased expression of calpain 6 in uterine sarcomas and carcinosarcomas: an immunohistochemical analysis. Int J Gynecol Cancer 2007;17:248-53. [Crossref] [PubMed]
- Lee SJ, Kim BG, Choi YL, et al. Increased expression of calpain 6 during the progression of uterine cervical neoplasia: immunohistochemical analysis. Oncol Rep 2008;19:859-63. [Crossref] [PubMed]
- Yoshikawa Y, Mukai H, Hino F, et al. Isolation of two novel genes, down-regulated in gastric cancer. Jpn J Cancer Res 2000;91:459-63. [Crossref] [PubMed]
- Carillo S, Pariat M, Steff AM, et al. Differential sensitivity of FOS and JUN family members to calpains. Oncogene 1994;9:1679-89. [PubMed]
- Kubbutat MH, Vousden KH. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol Cell Biol 1997;17:460-8. [Crossref] [PubMed]
- Zhou J, Köhl R, Herr B, et al. Calpain mediates a von Hippel-Lindau protein-independent destruction of hypoxia-inducible factor-1alpha. Mol Biol Cell 2006;17:1549-58. [Crossref] [PubMed]
- Gregoriou M, Willis AC, Pearson MA, et al. The calpain cleavage sites in the epidermal growth factor receptor kinase domain. Eur J Biochem 1994;223:455-64. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Winter JM, Brennan MF, Tang LH, et al. Survival after resection of pancreatic adenocarcinoma: results from a single institution over three decades. Ann Surg Oncol 2012;19:169-75. [Crossref] [PubMed]
- Zhai S, Huo Z, Wang Y, et al. TRIANGLE operation for borderline resectable pancreatic cancer in total pancreatectomy. Transl Cancer Res 2019;8:2416-24. [Crossref]
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371:1039-49. [Crossref] [PubMed]
- Sun J, Yan J, Qiao HY, et al. Loss of TRIM29 suppresses cancer stem cell-like characteristics of PDACs via accelerating ISG15 degradation. Oncogene 2020;39:546-59. [Crossref] [PubMed]
- Yu LM, Zhu YS, Xu CZ, et al. High calpain-1 expression predicts a poor clinical outcome and contributes to tumor progression in pancreatic cancer patients. Clin Transl Oncol 2019;21:924-32. [Crossref] [PubMed]
- Fong PY, Fesinmeyer MD, White E, et al. Association of diabetes susceptibility gene calpain-10 with pancreatic cancer among smokers. J Gastrointest Cancer 2010;41:203-8. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241. [Crossref] [PubMed]
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Rios-Doria J, Kuefer R, Ethier SP, et al. Cleavage of beta-catenin by calpain in prostate and mammary tumor cells. Cancer Res 2004;64:7237-40. [Crossref] [PubMed]
- Liu T, Mendes DE, Berkman CE. Prolonged androgen deprivation leads to overexpression of calpain 2: implications for prostate cancer progression. Int J Oncol 2014;44:467-72. [Crossref] [PubMed]
- Chen Y, Tang J, Lu T, et al. CAPN1 promotes malignant behavior and erlotinib resistance mediated by phosphorylation of c-Met and PIK3R2 via degrading PTPN1 in lung adenocarcinoma. Thorac Cancer 2020;11:1848-60. [Crossref] [PubMed]
- Meng XN, Jin Y, Yu Y, et al. Characterisation of fibronectin-mediated FAK signalling pathways in lung cancer cell migration and invasion. Br J Cancer 2009;101:327-34. [Crossref] [PubMed]
- Trillsch F, Kuerti S, Eulenburg C, et al. E-Cadherin fragments as potential mediators for peritoneal metastasis in advanced epithelial ovarian cancer. Br J Cancer 2016;114:213-20. [Crossref] [PubMed]
- Moretti D, Del Bello B, Cosci E, et al. Novel variants of muscle calpain 3 identified in human melanoma cells: cisplatin-induced changes in vitro and differential expression in melanocytic lesions. Carcinogenesis 2009;30:960-7. [Crossref] [PubMed]
- Moretti D, Del Bello B, Allavena G, et al. Calpain-3 impairs cell proliferation and stimulates oxidative stress-mediated cell death in melanoma cells. PLoS One 2015;10:e0117258. [Crossref] [PubMed]
- Tan Y, Wu C, De Veyra T, et al. Ubiquitous calpains promote both apoptosis and survival signals in response to different cell death stimuli. J Biol Chem 2006;281:17689-98. [Crossref] [PubMed]
- Bertoli C, Copetti T, Lam EW, et al. Calpain small-1 modulates Akt/FoxO3A signaling and apoptosis through PP2A. Oncogene 2009;28:721-33. [Crossref] [PubMed]
- Gu J, Xu FK, Zhao GY, et al. Capn4 promotes non-small cell lung cancer progression via upregulation of matrix metalloproteinase 2. Med Oncol 2015;32:51. [Crossref] [PubMed]
- Sharaf M, Antonios S, Mina S, et al. The scabicide effect of moxidectin in vitro and in experimental animals: Parasitological, histopathological and immunological evaluation. Exp Parasitol 2020;217:107961. [Crossref] [PubMed]
- Zheng PC, Chen X, Zhu HW, et al. Capn4 is a marker of poor clinical outcomes and promotes nasopharyngeal carcinoma metastasis via nuclear factor-κB-induced matrix metalloproteinase 2 expression. Cancer Sci 2014;105:630-8. [Crossref] [PubMed]
- Zhang C, Bai DS, Huang XY, et al. Prognostic significance of Capn4 overexpression in intrahepatic cholangiocarcinoma. PLoS One 2013;8:e54619. [Crossref] [PubMed]
- Dear N, Matena K, Vingron M, et al. A new subfamily of vertebrate calpains lacking a calmodulin-like domain: implications for calpain regulation and evolution. Genomics 1997;45:175-84. [Crossref] [PubMed]
- Schaefer KA, Toral MA, Velez G, et al. Calpain-5 Expression in the Retina Localizes to Photoreceptor Synapses. Invest Ophthalmol Vis Sci 2016;57:2509-21. [Crossref] [PubMed]
- Singh R, Brewer MK, Mashburn CB, et al. Calpain 5 is highly expressed in the central nervous system (CNS), carries dual nuclear localization signals, and is associated with nuclear promyelocytic leukemia protein bodies. J Biol Chem 2014;289:19383-94. [Crossref] [PubMed]
- Tonami K, Kurihara Y, Aburatani H, et al. Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol Cell Biol 2007;27:2548-61. [Crossref] [PubMed]
- Liu Y, Mei C, Sun L, et al. The PI3K-Akt pathway regulates calpain 6 expression, proliferation, and apoptosis. Cell Signal 2011;23:827-36. [Crossref] [PubMed]
- Skubitz KM, Skubitz AP. Differential gene expression in leiomyosarcoma. Cancer 2003;98:1029-38. [Crossref] [PubMed]
- Oh M, Rho SB, Son C, et al. Non-proteolytic calpain-6 interacts with VEGFA and promotes angiogenesis by increasing VEGF secretion. Sci Rep 2019;9:15771. [Crossref] [PubMed]
- Liu H, Jiang Y, Jin X, et al. CAPN 7 promotes the migration and invasion of human endometrial stromal cell by regulating matrix metalloproteinase 2 activity. Reprod Biol Endocrinol 2013;11:64. [Crossref] [PubMed]
- Al-Sinani S, Woodhouse N, Al-Mamari A, et al. Association of gene variants with susceptibility to type 2 diabetes among Omanis. World J Diabetes 2015;6:358-66. [Crossref] [PubMed]
- Yan ST, Li CL, Tian H, et al. Association of calpain-10 rs2975760 polymorphism with type 2 diabetes mellitus: a meta-analysis. Int J Clin Exp Med 2014;7:3800-7. [PubMed]
- Chan D, Tsoi MY, Liu CD, et al. Oncogene GAEC1 regulates CAPN10 expression which predicts survival in esophageal squamous cell carcinoma. World J Gastroenterol 2013;19:2772-80. [Crossref] [PubMed]
- Zha C, Farah CA, Holt RJ, et al. Biallelic variants in the small optic lobe calpain CAPN15 are associated with congenital eye anomalies, deafness and other neurodevelopmental deficits. Hum Mol Genet 2020;29:3054-63. [Crossref] [PubMed]
- Hata S, Doi N, Kitamura F, et al. Stomach-specific calpain, nCL-2/calpain 8, is active without calpain regulatory subunit and oligomerizes through C2-like domains. J Biol Chem 2007;282:27847-56. [Crossref] [PubMed]
- Hata S, Abe M, Suzuki H, et al. Calpain 8/nCL-2 and calpain 9/nCL-4 constitute an active protease complex, G-calpain, involved in gastric mucosal defense. PLoS Genet 2010;6:e1001040. [Crossref] [PubMed]
- Peng P, Wu W, Zhao J, et al. Decreased expression of Calpain-9 predicts unfavorable prognosis in patients with gastric cancer. Sci Rep 2016;6:29604. [Crossref] [PubMed]
- Dear TN, Möller A, Boehm T. CAPN11: A calpain with high mRNA levels in testis and located on chromosome 6. Genomics 1999;59:243-7. [Crossref] [PubMed]
- Macqueen DJ, Delbridge ML, Manthri S, et al. A newly classified vertebrate calpain protease, directly ancestral to CAPN1 and 2, episodically evolved a restricted physiological function in placental mammals. Mol Biol Evol 2010;27:1886-902. [Crossref] [PubMed]
- Bochner R, Samuelov L, Sarig O, et al. Calpain 12 Function Revealed through the Study of an Atypical Case of Autosomal Recessive Congenital Ichthyosis. J Invest Dermatol 2017;137:385-93. [Crossref] [PubMed]
- Dear TN, Meier NT, Hunn M, et al. Gene structure, chromosomal localization, and expression pattern of Capn12, a new member of the calpain large subunit gene family. Genomics 2000;68:152-60. [Crossref] [PubMed]
- Dear TN, Boehm T. Identification and characterization of two novel calpain large subunit genes. Gene 2001;274:245-52. [Crossref] [PubMed]
- Cherian JJ, Kapadia BH, Banerjee S, et al. Surgical intra-operative blood management strategies for total hip arthroplasty. Surg Technol Int 2014;24:319-25. [PubMed]
- Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539. [Crossref] [PubMed]
- Litosh VA, Rochman M, Rymer JK, et al. Calpain-14 and its association with eosinophilic esophagitis. J Allergy Clin Immunol 2017;139:1762-1771.e7. [Crossref] [PubMed]
- Pan H, Yu T, Sun L, et al. LncRNA FENDRR-mediated tumor suppression and tumor-immune microenvironment changes in non-small cell lung cancer. Transl Cancer Res 2020;9:3946-59. [Crossref]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010;140:883-99. [Crossref] [PubMed]
- Hiraoka N, Yamazaki-Itoh R, Ino Y, et al. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology 2011;140:310-21. [Crossref] [PubMed]
- Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004;10:942-9. [Crossref] [PubMed]
- Ino Y, Yamazaki-Itoh R, Shimada K, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer 2013;108:914-23. [Crossref] [PubMed]
- De Monte L, Reni M, Tassi E, et al. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J Exp Med 2011;208:469-78. [Crossref] [PubMed]
- Hollern DP, Xu N, Thennavan A, et al. B Cells and T Follicular Helper Cells Mediate Response to Checkpoint Inhibitors in High Mutation Burden Mouse Models of Breast Cancer. Cell 2019;179:1191-1206.e21. [Crossref] [PubMed]
- Gu-Trantien C, Migliori E, Buisseret L, et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight 2017;2:e91487. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)




