
Zebrafish xenograft model for studying the function of lncRNA SNHG4 in the proliferation and migration of colorectal cancer
Introduction
Zebrafish xenografts have developed into a promising experimental model for human cancer research (1-3). Many human cancer cell line-derived xenografts (CDX) have been successfully established in zebrafish, including breast cancer, lung cancer, melanoma, colorectal cancer (CRC), and gastric cancer (4-9). Additionally, a zebrafish patient-derived xenograft (zPDX) was recently developed, reflecting the tumor biology characteristics of patients (5,8). Compared to a mouse xenograft model, zebrafish xenografts have many unique advantages for cancer research, including the optical transparency of zebrafish larvae, which allows for easy in vivo observation, the proliferation and migration could be assessed simultaneously at 96 hours (10-12). Several studies have shown that the zebrafish xenograft model is a reliable tool for studying tumor growth and metastasis (11,13,14). Thus, the zebrafish xenograft model is a powerful and effective model for human cancer research.
CRC is one of the most commonly diagnosed cancers, causing a high number of cancer-related deaths worldwide (15). Although there have been advancements in early diagnosis and treatment for CRC patients, the prognosis is poor and the survival rate remains low (16-18). Therefore, there is an urgent need to better understand the fundamental mechanisms of CRC proliferation and migration in order to improve early diagnosis, treatment, and prognosis of CRC.
Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides with noncoding protein coding potential (19,20). Numerous studies have revealed that lncRNAs are involved in the progression of CRC, including proliferation, migration, invasion, and apoptosis (21-25). Thus far, zebrafish models have been used to investigate the oncogenic role of lncRNA THOR and LINC00152 in the progression of melanoma and lung cancer, respectively (26,27). Additionally, previous studies have reported the successful transplantation of human CRC cell lines into zebrafish to simultaneously evaluate proliferation and migration potential (5). These results suggested that zebrafish xenografts could be used to study the role of lncRNAs in CRC.
Small nucleolar RNA host genes (SNHGs) have been shown to play a role in the progression of multiple cancers, including CRC (28,29). For example, SNHG1 facilitated the CRC cell proliferation and migration (30), SNHG3 promoted growth of CRC cell (31). SNHG4, located on chromosome 5q31.2, is a proven oncogenic lncRNA in several types of cancers, including lung cancer and prostate cancer (27,32-35). Recently, a few of studies reported that SNHG4 was high expressed in CRC tissues and regulated the proliferation and migration of CRC cells in vitro, and it also could regulate the proliferation of CRC in mouse model (36,37). However, its roles of SNHG4 in CRC metastasis effect and underlying mechanisms of SNHG4 in CRC, a simple and fast in vivo model is required.. As zebrafish xenografts have been successfully used to investigate the function of lncRNAs in human cancers, especially metastasis, we chose zebrafish CRC xenograft model to verify the function of SNHG4 in CRC.
In this study, we used Gene Expression Profiling Interactive Analysis (GEPIA) software and found that SNHG4 was upregulated in CRC. Meanwhile, we detected the expression level of SNHG4 in CRC cell lines. We then investigated the effect on proliferation and migration of SNHG4 knockdown in vitro and in zebrafish xenografts in vivo, respectively. In addition, we detected epithelial mesenchymal transition (EMT)-related genes after knockdown of SNHG4 in CRC cells. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-832/rc).
Methods
Zebrafish husbandry
Adult zebrafish were maintained in an automated fish culture system (Haisheng, China) at 28 °C and a 14-hour light/10-hour dark cycle. Embryos were cultured in 10% Hank’s solution consisting of (in mM) 140 NaCl, 5.4 KCl, 0.25 Na2HPO4, 0.44 KH2PO4, 1.3 CaCl2, 1.0 MgSO4, and 4.2 NaHCO3 (pH 7.2). In our study, 48-hour post-fertilization (hpf) wild-type AB or Tg(fli1a:EGFP) zebrafish were used in all experiments (38). After injection, the zebrafish were cultured at 34 °C. Experiments were performed under a project license (No. HEYLL201935) granted by Affiliated Huai’an Hospital of Xuzhou Medical University, in compliance with the Affiliated Huai’an Hospital of Xuzhou Medical University institutional guidelines for the care and use of animals.
Gene expression analysis and cell culture
The expression level of lncRNA SNHG4 in normal tissue samples compared to tumor samples was analyzed using the GEPIA database (https://gepia.cancer-pku.cn/), which contains 275 colon adenocarcinoma tissue samples and 41 normal tissue samples, and 92 rectum adenocarcinoma tissue samples and 10 normal tissue samples. Human CRC cell lines were obtained from the Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (Shanghai, China) and included SW480, HCT116, HT29, LoVo, and normal colon epithelial cells [fetal human colon (FHC)]. HCT116 and FHC cells were cultured in 1640 medium, HT29 cells were grown in McCoy’s 5a medium, and LoVo cells were cultured in F-12k medium. All media were supplemented with 10% FBS (fetal bovine serum), 100 U/mL penicillin, and 100 µg/mL streptomycin. Cells were placed in a humidified atmosphere with 5% CO2 at 37 °C. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was reverse transcribed to complementary DNA (cDNA) using random primers according to the instructions of the 1st Strand cDNA Synthesis SuperMix for qPCR kit (Takara, Dalian, China). Quantitative real-time PCR analysis was performed to detect the expression level of SNHG4 in the different CRC cell lines and normal colon epithelial cells using SYBR Green Master Mix kit (Takara) following the manufacturer’s protocol. Data were collected and analyzed based on the 2-∆∆CT method and normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The specific primer sequences of SNHG4 were as follows: forward 5'-gcagatgcaactgcaggatgac-3' and reverse 5'-tgtcattcaggtgaaggcatctg-3'. The primer sequences for GAPDH were as follows: forward 5'-GGGAGCCAAAAGGGTCAT-3' and reverse 5'-GAGTCCTTCCACGATACCAA-3'.
RNA interference
Small interfering RNA (siRNA) targeting SNHG4 and negative control were purchased from General Biosystems (Beijing, China). The cells were seeded in 6-well plates and after 24 hours, the siRNAs were transfected into the cells using Lipofectamine 2000 reagents (Invitrogen) according to the manufacturer’s protocol and the concentration of siRNA was 100 nM. Silencing efficiency of the siRNAs was analyzed by qRT-PCR 24 hours after transfection with siRNAs. The SNHG4 siRNA (si-SNHG4) and negative control siRNA (NC) sequences were 5'-gtgacaccaagataagtaa-3' and 5'-TTCTCCGAACGTGTCACGT-3', respectively.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay were performed to analyze the cell proliferation using CCK-8 (DOJINDO, Japan). A total of 2×103 cells transfected with si-SNHG4 negative control siRNA and untransfected cells (normal control) were seed in 96-well plates. 10 μL of the CCK-8 reagent was added to each well at the same time, which containing 100 μL culture medium. After 2 h, the product was measured the optical density (OD) at 450 nm by monitoring on a microplate reader (BioTek Elx800, USA) according to the manufacturer’s instructions. Every 24 h was detected from 0 to 96 h.
Colony formation assay
Cell proliferation was measured using the colony formation assay. Cells were seeded in 6-well plates and transfected with siRNAs. After 24 hours, a total of 50 cells transfected with siRNAs were seeded per well in 6-well plates and cultured in a medium containing 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. The medium was replaced every 3 days. After 14 days, the colonies were fixed with methanol for 30 minutes and stained with 0.1% crystal violet for 30 minutes. The visible colonies were photographed by digital camera and counted.
Transwell assay
The transfected cells were seeded in 24-well plates with 8-mm-pore size chamber inserts. Cells (10×104) diluted with 200 μL serum-free culture medium were plated into the upper chambers of the 24-well plates, and 800 μL medium with 10% FBS was placed in the lower chambers. After 36 hours, the cells that had moved from the upper membrane to the bottom surface of the membrane were treated with methanol for 30 minutes and stained with 0.1% crystal violet for 30 minutes. The images were acquired under a microscope (10×).
Zebrafish xenograft injection and imaging in vivo and quantitative analysis
The cells were plated into the six-plates, after cultured 24 hours, NC or si-SNHG4 were transfected. After transfection 24 hours later, the cells were collected and labeled with CM-DiI (Invitrogen) before injection. The cultured cells were treated as previously described (39). We then injected approximately 300 CM-DiI labeled cells into the perivitelline space (PVS) of the 48-hpf zebrafish larvae, which were fixed with 1.2% low-melting gel (Promega Corp., Madison, WI, USA) under a Picospritzer III microinjector (Parker-Hannifin Corp., Cleveland, OH, USA). After injection, the zebrafish larvae were cultured at 34 °C. After 24 hours post-injection (hpi), similar sizes of transplanted cells were selected for further research and the zebrafish were maintained at 34 °C until the experiment’s end.
At 4 days post-injection (dpi), images of the zebrafish larvae, which were fixed with 1.2% low-melting gel, were acquired via stereo microscopy (MVX10, Olympus, Tokyo, Japan) or confocal microscopy using a 20× water-immersion objective (Fluoview 1000, Olympus). The image resolution was 1,600×1,200 (MVX10) or 1,024×1,024 (Fluoview 1000) pixels.
Detection of related genes after silencing SNHG4
A previous study found that SNHG4 knockdown inhibited migration by participating in the EMT process in lung cancer (32). After silencing SNHG4 by transfecting cells with si-SNHG4, we used qPCR to detect N-cadherin, E-cadherin, Vimentin, matrix metalloproteinase-2 (MMP2), MMP9, and mitogen-activated protein kinase 1 (MAPK1) messenger RNA (mRNA) levels. The sequences of specific primers are listed in Table 1.
Table 1
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| N-cadherin | GCCCCTCAAGTGTTACCTCAA | AGCCGAGTGATGGTCCAATTT |
| E-cadherin | ACAATGCCGCCATCGCTTA | TGAACCACCAGGGTATACGTAGG |
| Vimentin | AAGTTTGCTGACCTCTCTGAGGCT | CTTCCATTTCACGCATCTGGCGTT |
| MMP2 | ATGCCGTCGTGGACCTGC | TGCTTCCAAACTTCACGCTCTT |
| MMP9 | GCCTTCGCACTGTGGAGC | GGATACCCGTCTCCGTGCTC |
| MAPK1 | GACCTCAAGCCTTCCAACCTG | TGGAGCCCTGTACCAACGTG |
PCR, polymerase chain reaction.
Statistical analysis
All statistical data were analyzed using unpaired Student’s t-tests. A value of P<0.05 was considered statistically significant. All results are represented as the mean ± standard error of mean (SEM).
Results
LncRNA SNHG4 is upregulated in CRC
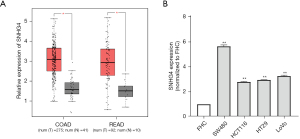
We used GEPIA software to analyze the database and found that the expression of lncRNA SNHG4 was relatively high in CRC tissues (Figure 1A). We then detected SNHG4 expression levels in 4 human CRC cell lines (SW480, HCT116, HT29, and LoVo) and normal colon epithelial cells (FHC). We found that SNHG4 expression was higher in the SW480, HCT116, HT29, and LoVo cell lines (fold values were 5.66, 2.79, 2.95, and 3.26, respectively) than normal colon epithelial cells (Figure 1B), which was consistent with the results of GEPIA database analysis.
Silencing SNHG4 inhibited the proliferation and migration of CRC cells in vitro
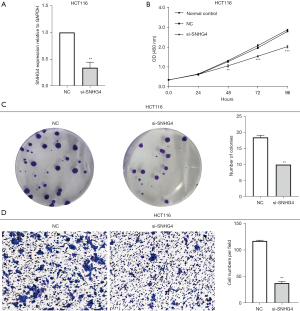
To investigate the function of SNHG4 in CRC cells, we silenced its expression by transfecting cells with siRNA. In accordance with the reported data on siRNA targeting SNHG4 (27), we synthesized the si-SNHG4 and NC and then transfected siRNAs into CRC cell lines. After 24 hours, silencing efficiency was detected by qRT-PCR. The results showed that compared with NC, silencing efficiency in the HCT116 cell line of SNHG4 was 65.8%, which was higher than the SW480 (42.7%) and LoVo (24.7%) cell lines (Figure 2A), and thus we chose to use HCT116 cells for further investigations. The CCK-8 assay indicated that the proliferation was inhibited (Figure 2B) and colony formation assay showed that colony formation ability was significantly reduced after knocking down SNHG4 (Figure 2C). And we found the untransfected group had no difference with the transfecting NC siRNA group, also. So the normal control group was not added in subsequent experiments in vitro. We then performed the transwell assay to detect the effect of SNHG4 on HCT116 cell migration. The results showed that compared with the NC group, cell migration was obviously repressed in HCT116 cells after silencing SNHG4 (Figure 2D).
These results demonstrated that SNHG4 played an oncogenic role in CRC cells, which prompted us to perform further verification of the functional phenotype in vivo in a zebrafish xenograft model via siRNA-mediated knockdown.
Silencing SNHG4 inhibited the proliferation and migration of CRC cells in the zebrafish xenograft model based on stereo microscopy and confocal microscopy
We performed in vivo tests in the zebrafish xenograft model to verify whether silencing SNHG4 by siRNA affected the function of CRC by transplanting HCT116 cells transfected with si-SNHG4 into zebrafish embryos. Cells transfected with si-SNHG4 or NC were labeled with fluorescent dye (CM-DiI), and approximately 300 cells were then injected into the PVS of 48-hpf zebrafish larvae. At 1 dpi, we collected similar-sized fluorescent areas from the transplanted zebrafish larvae via fluorescence stereo microscopy. At 4 dpi, the images were acquired, which included the yolk and trunk of the zebrafish. The CM-DiI positive signals of the zebrafish yolk and trunk were quantified, with the tumor area of the yolk and trunk representing the proliferation and migration of cells, respectively (5,39). The bright field image of zebrafish at 4 dpi was photographed through stereo microscopy (Figure 3A). The results showed that the CM-DiI positive area of the silenced-SNHG4 yolk and trunk was obviously smaller than the NC group (Figure 3B,3C), demonstrating that proliferation and migration were decreased after silencing SNHG4 in HCT116 cells (Figure 3D,3E). The results of the zebrafish xenograft model were consistent with the data from the in vitro experiments, verifying the oncogenic role of SNHG4 in CRC.
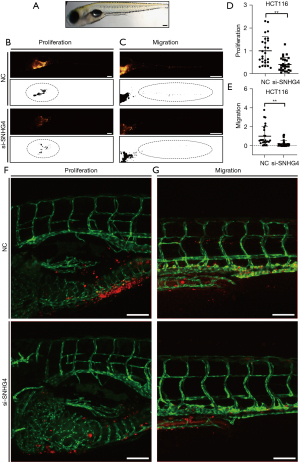
In addition, we used confocal microscopy to exclude the few nonspecific signals and acquire high-resolution images. In this test, we injected the CM-DiI labeled cells into Tg(fli1a:EGFP) zebrafish larvae, which contain enhanced green fluorescent protein (EGFP)-labeled vascular endothelial cells, and we simultaneously observed angiogenesis. At 4 dpi, we imaged the xenografts using multilayer scanning (Figure 3F,3G). By analyzing the confocal images, we found cell proliferation and migration significantly decreased after silencing SNHG4, although a few noncell spots existed in CM-DiI-positive signals in the yolk and trunk compared with the stereo microscopy images.
These results verified the functional phenotype after silencing SNHG4 in vivo in a zebrafish xenograft model, demonstrating that this model could be used to research the function of SNHG4 in CRC.
Detection of EMT-related genes after silencing SNHG4
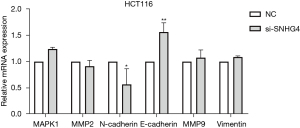
After silencing SNHG4 in HCT116 cells, we detected mRNA levels of EMT-related genes by qPCR. These genes contained N-cadherin, E-cadherin, Vimentin, MMP2, MMP9, MAPK1. Compared to the NC group, the mRNA level of E-cadherin was significantly upregulated and N-cadherin was obviously downregulated in the SNHG4-silenced group, while there was no significant difference in the mRNA levels of Vimentin, MMP2, MMP9, and MAPK1 (Figure 4). These results indicated that SNHG4 participated in migration in HCT116 cells by regulating EMT-related genes.
Discussion
In this study, we found SNHG4 was upregulated in CRC tissues and cell lines. Moreover, cell proliferation and migration were suppressed after silencing SNHG4 in vitro. Meanwhile, the functional phenotype was verified in zebrafish xenografts using stereo and confocal microscopy and was consistent with the in vitro results. These data showed that SNHG4 played an oncogenic role in CRC and strongly suggested that zebrafish xenograft was a reliable model for functional research of lncRNAs in human cancer. In addition, we detected EMT-related genes after silencing SNHG4, which upregulated E-cadherin and downregulated N-cadherin in HCT116 cells. These results demonstrated that SNHG4 knockdown inhibited EMT, which contributed to our understanding of the underlying mechanism of SNHG4 in CRC.
Numerous recent studies have indicated that lncRNAs play a crucial role in the development and progression of human cancer. In CRC, the biological functions of several lncRNAs have been reported, including GAS5, LINRIS, GLCC1, and NEAT1 (40-43). The role of SNHG4 has been investigated in many types of cancers, such as osteosarcoma, lung cancer, prostate cancer, glioblastoma, and cervical cancer, and SNHG4 has been reported to be involved in human cancer progression, including proliferation, migration, invasion, and apoptosis (27,32-34,44). In lung cancer and neuroblastoma, silencing SNHG4 suppressed EMT to inhibit tumor metastasis (32,35). EMT is a complicated cellular process, which the epithelial cells acquired a mesenchymal phenotype and plays an important role in the development of tumor metastasis. Some lncRNAs have been shown to regulate EMT-related genes involved in the progression of CRC (45). In our study, we similarly detected EMT-related genes after silencing SNHG4 in CRC, suggesting that SNHG4 might be involved in the progression of CRC by EMT. These results further contributed to our understanding of the underlying mechanisms of SNHG4 in CRC.
In our study, we used zebrafish xenografts as an in vivo tool to investigate the role of SNHG4 in the proliferation and migration of CRC cells. Compared with mouse models, zebrafish xenografts have unique advantages. First, a zebrafish xenograft offers speed in tumor biology studies. The proliferation and migration of tumor cells can be evaluated 4 days after transplantation, while mouse xenografts require 2–4 weeks and must be transfected with short hairpin RNA (shRNA). Second, after transplantation of tumor cells, growth and migration can be visualized simultaneously in vivo in zebrafish larvae (46). Further, the development of tumor cells at early stages can also be monitored in vivo, which is not possible in mouse models. Third, due to the transparent nature of zebrafish larvae, tumor cell behavior can be detected at the single-cell level in zebrafish xenografts in vivo. With the rapid development of imaging technology, including high-resolution, timely, and 3-dimensional (3D) imaging, the interaction of different cells in the tumor microenvironment or the interaction between tumor cells and other cells can be observed and imaged in vivo (6). Lastly, the operating procedures of using a zebrafish model is relatively simple compared to a mouse model. Cells are injected into the PVS of zebrafish larvae, while tail vein injection, which is a more difficult procedure, is performed in a mouse model (47).
On the basis of these advantages, zebrafish xenografts have been used for drug screening in vivo, including cetuximab and regorafenib for CRC therapy. Cetuximab was found to induce apoptosis of xenografts after Hke3 and HCT116 cells were transplanted into zebrafish, and regorafenib induced apoptosis and antiangiogenic activity after transplantation of HT29 cells (5). The effects of cetuximab and regorafenib could be observed in zebrafish in vivo in a short time, suggesting that the zebrafish xenograft model is an effective and fast in vivo model for drug testing in human cancer.
In addition, human CRC cells have been successfully transplanted into zebrafish, with blood vessels observed at 4 dpi (5). In our study, we used Tg(fli1a:EGFP) zebrafish larvae to evaluate vascular change in zebrafish xenografts after transplanting CRC cells. Due to insufficient accurate data, we did not observe obvious blood vessel changes. However, these data suggested that a zebrafish model could be used to research the interactions between the tumor cells and the microenvironment. These findings strongly indicated that the zebrafish xenograft is a reliable and promising model for human cancer research.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-832/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-832/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-832/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. HEYLL201935) granted by Affiliated Huai’an Hospital of Xuzhou Medical University, in compliance with the Affiliated Huai’an Hospital of Xuzhou Medical University institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet 2007;8:353-67. [Crossref] [PubMed]
- Snaar-Jagalska BE. ZF-CANCER: developing high-throughput bioassays for human cancers in zebrafish. Zebrafish 2009;6:441-3. [Crossref] [PubMed]
- Hason M, Bartůněk P. Zebrafish Models of Cancer-New Insights on Modeling Human Cancer in a Non-Mammalian Vertebrate. Genes (Basel) 2019;10:935. [Crossref] [PubMed]
- Teng Y, Xie X, Walker S, et al. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer 2013;13:453. [Crossref] [PubMed]
- Fior R, Póvoa V, Mendes RV, et al. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc Natl Acad Sci U S A 2017;114:E8234-43. [Crossref] [PubMed]
- Liu C, Zhang Y, Lim S, et al. A Zebrafish Model Discovers a Novel Mechanism of Stromal Fibroblast-Mediated Cancer Metastasis. Clin Cancer Res 2017;23:4769-79. [Crossref] [PubMed]
- Lu JW, Ho YJ, Ciou SC, et al. Innovative Disease Model: Zebrafish as an In Vivo Platform for Intestinal Disorder and Tumors. Biomedicines 2017;5:58. [Crossref] [PubMed]
- Wu JQ, Zhai J, Li CY, et al. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in gastric cancer. J Exp Clin Cancer Res 2017;36:160. [Crossref] [PubMed]
- Fornabaio G, Barnhill RL, Lugassy C, et al. Angiotropism and extravascular migratory metastasis in cutaneous and uveal melanoma progression in a zebrafish model. Sci Rep 2018;8:10448. [Crossref] [PubMed]
- Lee LM, Seftor EA, Bonde G, et al. The fate of human malignant melanoma cells transplanted into zebrafish embryos: assessment of migration and cell division in the absence of tumor formation. Dev Dyn 2005;233:1560-70. [Crossref] [PubMed]
- Marques IJ, Weiss FU, Vlecken DH, et al. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer 2009;9:128. [Crossref] [PubMed]
- Wertman J, Veinotte CJ, Dellaire G, et al. The Zebrafish Xenograft Platform: Evolution of a Novel Cancer Model and Preclinical Screening Tool. Adv Exp Med Biol 2016;916:289-314. [Crossref] [PubMed]
- Haldi M, Ton C, Seng WL, et al. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis 2006;9:139-51. [Crossref] [PubMed]
- Stoletov K, Montel V, Lester RD, et al. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc Natl Acad Sci U S A 2007;104:17406-11. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Villeneuve PJ, Sundaresan RS. Surgical management of colorectal lung metastasis. Clin Colon Rectal Surg 2009;22:233-41. [Crossref] [PubMed]
- Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017;170:548-563.e16. [Crossref] [PubMed]
- Rychahou P, Bae Y, Reichel D, et al. Colorectal cancer lung metastasis treatment with polymer-drug nanoparticles. J Control Release 2018;275:85-91. [Crossref] [PubMed]
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559-63. [Crossref] [PubMed]
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell 2011;145:178-81. [Crossref] [PubMed]
- Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer 2017;16:9. [Crossref] [PubMed]
- Shan Y, Ma J, Pan Y, et al. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis 2018;9:722. [Crossref] [PubMed]
- Sun J, Hu J, Wang G, et al. LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer. J Exp Clin Cancer Res 2018;37:106. [Crossref] [PubMed]
- Liang ZX, Liu HS, Wang FW, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis 2019;10:829. [Crossref] [PubMed]
- Xu M, Xu X, Pan B, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer 2019;18:135. [Crossref] [PubMed]
- Hosono Y, Niknafs YS, Prensner JR, et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 2017;171:1559-1572.e20. [Crossref] [PubMed]
- Xu R, Feng F, Yu X, et al. LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma. Cell Prolif 2018;51:e12515. [Crossref] [PubMed]
- Abel Y, Rederstorff M. SnoRNAs and the emerging class of sdRNAs: Multifaceted players in oncogenesis. Biochimie 2019;164:17-21. [Crossref] [PubMed]
- Yang H, Jiang Z, Wang S, et al. Long non-coding small nucleolar RNA host genes in digestive cancers. Cancer Med 2019;8:7693-704. [Crossref] [PubMed]
- Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget 2017;9:2128-39. [Crossref] [PubMed]
- Huang W, Tian Y, Dong S, et al. The long non-coding RNA SNHG3 functions as a competing endogenous RNA to promote malignant development of colorectal cancer. Oncol Rep 2017;38:1402-10. [Crossref] [PubMed]
- Tang Y, Wu L, Zhao M, et al. LncRNA SNHG4 promotes the proliferation, migration, invasiveness, and epithelial-mesenchymal transition of lung cancer cells by regulating miR-98-5p. Biochem Cell Biol 2019;97:767-76. [Crossref] [PubMed]
- Wang X, Tian W, Wu L, et al. LncRNA SNHG4 regulates miR-138/c-Met axis to promote the proliferation of glioblastoma cells. Neuroreport 2020;31:657-62. [Crossref] [PubMed]
- Wang ZY, Duan Y, Wang P. SP1-mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR-377 to facilitate prostate cancer progression through regulation of ZIC5. J Cell Physiol 2020;235:3916-27. [Crossref] [PubMed]
- Yang H, Guo JF, Zhang ML, et al. LncRNA SNHG4 promotes neuroblastoma proliferation, migration, and invasion by sponging miR-377-3p. Neoplasma 2020;67:1054-62. [Crossref] [PubMed]
- Zhou N, Chen Y, Yang L, et al. LncRNA SNHG4 promotes malignant biological behaviors and immune escape of colorectal cancer cells by regulating the miR-144-3p/MET axis. Am J Transl Res 2021;13:11144-61. [PubMed]
- Zhou Z, Tan F, Pei Q, et al. lncRNA SNHG4 modulates colorectal cancer cell cycle and cell proliferation through regulating miR-590-3p/CDK1 axis. Aging (Albany NY) 2021;13:9838-58. [Crossref] [PubMed]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 2002;248:307-18. [Crossref] [PubMed]
- Shen W, Pu J, Sun J, et al. Zebrafish xenograft model of human lung cancer for studying the function of LINC00152 in cell proliferation and invasion. Cancer Cell Int 2020;20:376. [Crossref] [PubMed]
- Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol 2018;11:113. [Crossref] [PubMed]
- Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer 2019;18:143. [Crossref] [PubMed]
- Tang J, Yan T, Bao Y, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun 2019;10:3499. [Crossref] [PubMed]
- Wang Y, Lu JH, Wu QN, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer 2019;18:174. [Crossref] [PubMed]
- Li H, Hong J, Wijayakulathilaka WSMA. Long non-coding RNA SNHG4 promotes cervical cancer progression through regulating c-Met via targeting miR-148a-3p. Cell Cycle 2019;18:3313-24. [Crossref] [PubMed]
- O'Brien SJ, Bishop C, Hallion J, et al. Long non-coding RNA (lncRNA) and epithelial-mesenchymal transition (EMT) in colorectal cancer: a systematic review. Cancer Biol Ther 2020;21:769-81. [Crossref] [PubMed]
- Zhao C, Wang X, Zhao Y, et al. A novel xenograft model in zebrafish for high-resolution investigating dynamics of neovascularization in tumors. PLoS One 2011;6:e21768. [Crossref] [PubMed]
- Steel CD, Stephens AL, Hahto SM, et al. Comparison of the lateral tail vein and the retro-orbital venous sinus as routes of intravenous drug delivery in a transgenic mouse model. Lab Anim (NY) 2008;37:26-32. [Crossref] [PubMed]
(English Language Editor: A. Muijlwijk)


