
Mutational characteristics of young and elderly gastric cancer: a comparative study
Introduction
Gastric cancer (GC) is a common malignant cancer worldwide, and is especially prevalent in Asia regions including China, Japan, and Korea. Young gastric cancer (YGC, under 40 years old) has shown a poorer rate of diagnosis than elderly gastric cancer (EGC, above 70 years old). Clinically, YGC is associated with delayed diagnosis and being more aggressive. Generally, GC is considered to be associated with age, with its incidence peaking among those older than 50 years (1). In the last decades, characteristics of this neoplasm in young adults had been reported (2-6). The proportion of YGCs has varied from 6% to 8% (5-8). Most comparative studies of the clinicopathological characteristics between YGC and EGC have been conducted in Japan (9-12), and the different features of GCs between YGCs and EGCs has been demonstrated in a Japanese study (11).
It has been reported that YGC and EGC patients show different genomic profiles (13). Molecularly targeted therapy targeting sensitizing mutations has been a successful strategy for the clinical treatment of cancer (14). For example, the presence of epidermal growth factor receptor (EGFR)-positive mutations in lung cancer patients is the gold-standard biomarker for the first-line EGFR tyrosine kinase inhibitor (TKI) therapy. The rate of EGFR mutations is high in the Chinese cancer population (15,16). The purpose of the present study was to analyze the mutational landscape of GC, compare the molecular features of young patients with those of elderly patients, and to profile the point mutation frequency of 50 GC-associated genes using targeted next-generation sequencing, a more sensitive mutation detection technology.
Although some articles have reported the gene mutation characteristics of gastric cancer in the TCGA database, they mainly compared the relationship between the ACRG classification and the TCGA classification in evaluating the prognosis. In this paper, the mutation status and 50 cancer-related gene signatures of YGC (n=18) and EGC (n=18) patients in the TCGA-STAD database were classified and analyzed. The two have different focuses. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-934/rc).
Methods
Patient collection
A total of 8 GC patients were recruited from the Zhongshan Hospital of Xiamen University. Of these, 4 participants (50%) were 40 years of age or younger. This study was approved by the Ethics Committee of Zhongshan Hospital of Xiamen University (No. MULAC20180085), and performed in accordance with the principles of the Declaration of Helsinki (as revised in 2013). After the patients had provided informed consent to participate in the study, tumor tissues and their matched control samples were obtained for targeted next-generation sequencing. All samples were subjected to pathology review for histological subtyping and detailed clinical characteristics. The clinical characteristics of the 8 GCs are shown in Table 1.
Table 1
| Patient | Gender | Type | Age (years) |
|---|---|---|---|
| 1 | Male | Diffuse | 78 |
| 2 | Female | Diffuse | 77 |
| 3 | Female | Diffuse | 73 |
| 4 | Male | Diffuse | 75 |
| 5 | Female | Diffuse | 33 |
| 6 | Male | Diffuse | 19 |
| 7 | Male | Diffuse | 39 |
| 8 | Male | Diffuse | 34 |
PCR and barcoding
The 50 cancer-related genes included well-known oncogenes and tumor suppressor genes [as listed in the Candidate Cancer Gene Database:
Sequencing and data analysis
The barcoded PCR products were mixed and cleaned using AMPure beads (Beckman Coulter, Brea, CA, USA). The pooled amplicons were then mixed 7:3 with PhiX (Illumina, San Diego, CA, USA), denatured and clustered at 6 pM on Illumina Miseq 500 cycle flow-cell and sequenced (250 cycles forward, 10 cycles barcode, 250 cycles reverse). Raw trace files were processed with trimmomatic (version 0.36) (18) in paired-end mode to remove adapter sequences, and to filter out pairs with a sequence <100 nt to exclude short read artefacts. Local alignments of reads to the hg19 genome were performed using Burrows-Wheeler Aligner (BWA; version 0.7.17) (19) in paired-end mode. Sequence alignment map (SAM) files were converted to binary alignment map (BAM) files, sorted, and indexed using samtools (version 1.4) (20). Mutations were called using samtools/bcftools to generate the vcf files.
TCGA WES somatic mutations
Confident somatic mutation calls derived from the whole-exome sequencing (WES) data of the stomach adenocarcinoma (STAD) cohorts were directly downloaded from the Cancer Genome Atlas Genomic Data Commons (TCGA GDC) data portal using R/Bioconductor package ‘TCGAbiolinks’ (21). The somatic mutations were annotated with oncotator (22) using the same settings as in our analysis.
Statistical analysis
The R/maftools package was used to describe the mutational characteristics. Mutations were called using samtools/bcftools to generate the vcf files. The somatic mutations were annotated with oncotator using the same settings as in our analysis.
Results
The mutational landscape of YGC and EGC based on TCGA-STAD database
To declare the molecular difference between YGC and EGC groups, a distribution of gene aberrances, stratified by subgroups in Figure 1 was made based on TCGA-STAD database. The 40 patients were equally divided into young (n=20) and old (n=20) according to their age, forming a cohort to be analyzed. The mean age of the YGC group was 42.5 years old (30–46 years) and that of the EGC was 86.2 years old (83–90 years). Tumor mutation burden (TMB) was shown to be associated with clinical benefit of anti-programmed cell death protein 1 (PD-1) therapy. From the results, we observed that the EGC group harbored higher TMB level than the YGC group. As an oncogenic gene, TP53 showed the same mutation rate in YGC (4/18) and EGC (4/18). Most genes (19 out of 20) showed more somatic variation in the EGC group than the YGC group.

The comparation of 50 cancer-related genes in YGC and EGC form TCGA-STAD
In this study, the 50 cancer-related genes included well-known oncogenes and tumor suppressor genes [as listed in the Candidate Cancer Gene Database (23), such as CCND1, CCNE1, CDK6, CDKN2A, EGFR, ERBB2, FGFR2, KRAS, MET, MYC, and PTEN. Genetic alterations involving the phosphatidylinositol-3 kinase/AKT signaling pathway also occur in GCs, particularly in advanced and dedifferentiating tumors. The distribution of gene aberrances, stratified by subgroups in Figure 2 was made based on TCGA-STAD database. The EGC subgroup (n=20) harbored 35 missense mutations, and only in ALK, FBXW7, GNA11, PTPN11, and FGFR3; the YGC subgroup (n=20) had 23 missense mutations, and only in CDH1, FGFR2, CTNNB1, and ATM. Taken together, these data indicated that there is no obvious somatic mutations between YGC and EGC from TCGA-STAD database.
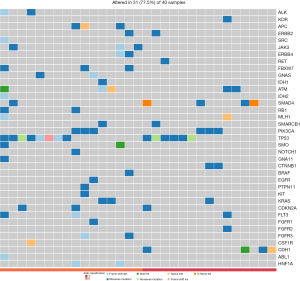
Targeted next-generation sequencing for mutations detection of 50 cancer-related genes in a Chinese cohort
The next-generation sequencing (NGS) offers single nucleotide level information on a different scale including whole-genome sequencing (WGS), WES, whole-transcriptome sequencing (WTS), and PCR-based targeting sequencing of multiple specific genomic regions. Whereas largescale analyses are essential for discovery projects, targeted panels with a well-known gene list may offer further advances in the routine molecular diagnostics of cancer. We collected 8 GC samples, as shown in Table 1, as our cohort for comparative study of GCs. In this comparative study, such an approach may be helpful to expand the currently existing 50 cancer-related gene panels to enable simultaneous testing for multiple mutations. The distribution of gene aberrances, stratified by the subgroups shown in Figure 3, was made based on our cohort. The EGC subgroup (n=4, >70 years old) harbored 11 missense mutations, and in ALK, FBXW7, GNA11, PTPN11, and FGFR3; the YGC subgroup (n=4, <40 years old) had 13 missense mutations, and only in CDH1, FGFR2, CTNNB1, and ATM. These alterations will affect many genes, and these affected genes will become the main carcinogens, including several therapeutic targets, such as ERBB2, FGFR2, and MET.

Discussion
A controversial issue emerged when the prognosis of YGC patients was mentioned, and it is generally believed that the EGC population has a better prognosis (11). However, it has been reported that compared with EGC, YGC has a better prognosis, which may be related to the better overall physical condition of young people. After all, the prognosis of the elderly may be affected by other common diseases of the elderly. In this comparative study, we find that the YGC group harbored more variation in 50 cancer-related genes than EGC using targeted NGS, while not in the TCGA-STAD database using WES; this may be the result of more reads and the depth of targeted NGS compared to WES. The detection of some mutations in YGC will further indicate the clinical benefit of YGC using target therapy. Indeed, due to the inclusion of additional mutations, such as those of KRAS and FGFR2, more YGCs were identified by the 50 cancer-related genes panel to carry at least one-point mutation.
This study had some limitations. First, the sample size of the research cohort was small and, therefore, the results should be interpreted carefully. Second, our cohort had no objective response or even progression-free survival analysis for molecularly targeted therapy, which increased the bias of the study. Third, the frequency of other 50 cancer-related genes mutations were not explored, which meant that some useful information was missing. Finally, only patients who was young or elderly were involved in this study and its analysis, leading to population bias.
Conclusions
In summary, by using PCR-based targeted NGS mutation detection, the present study demonstrated a higher proportion of 50 cancer-related genes mutation in YGC participants than EGC participants in our cohort. Through investigation of TMB's role in the predication of clinical response of checkpoint inhibitor therapy, a higher TMB level was revealed in EGC than YGC patients from TCGA-STAD database. The comparation of mutational characteristics in YGC and EGC will deepen the understanding of GC development and therapy for precision medicine.
Acknowledgments
We would also like to thank Shanghai Tongshu Biotechnology Co., Ltd. for their technical support.
Funding: This work was supported by grants from The National Natural Science Foundation of China (Nos. 81272445 and 81870388); Special Project for Marine Economic Development of Xiamen City (No. 17GYY001NF01).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-934/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-934/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-934/coif). All authors report they received technical support from Shanghai Tongshu Biotechnology Co., Ltd. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Zhongshan Hospital of Xiamen University (No. MULAC20180085), and performed in accordance with the principles of the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maehara Y, Sakaguchi Y, Moriguchi S, et al. Signet ring cell carcinoma of the stomach. Cancer 1992;69:1645-50. [Crossref] [PubMed]
- Kubicki S, Strojecki L. Cancer of the stomach in young adults. Pol Tyg Lek (Wars) 1957;12:1846-52. [PubMed]
- BLOCK M. GRIEP AH, POLLARD HM. The occurrence of gastric neoplasms in youth. Am J Med Sci 1948;215:398-404. [Crossref] [PubMed]
- BELLEGIE NJ. DAHLIN DC. Malignant disease of the stomach in young adults. Ann Surg 1953;138:7-12. [Crossref] [PubMed]
- TAMURA PY. CURTISS C. Carcinoma of the stomach in the young adult. Cancer 1960;13:379-85. [Crossref] [PubMed]
- Matley PJ, Dent DM, Madden MV, et al. Gastric carcinoma in young adults. Ann Surg 1988;208:593-6. [Crossref] [PubMed]
- Bloss RS, Miller TA, Copeland EM 3rd. Carcinoma of the stomach in the young adult. Surg Gynecol Obstet 1980;150:883-6. [PubMed]
- Tso PL, Bringaze WL 3rd, Dauterive AH, et al. Gastric carcinoma in the young. Cancer 1987;59:1362-5. [Crossref] [PubMed]
- Mitsudomi T, Matsusaka T, Wakasugi K, et al. A clinicopathological study of gastric cancer with special reference to age of the patients: an analysis of 1,630 cases. World J Surg 1989;13:225-30; discussion 230-1. [Crossref] [PubMed]
- Fujimoto S, Takahashi M, Ohkubo H, et al. Comparative clinicopathologic features of early gastric cancer in young and older patients. Surgery 1994;115:516-20. [PubMed]
- Maehara Y, Emi Y, Tomisaki S, et al. Age-related characteristics of gastric carcinoma in young and elderly patients. Cancer 1996;77:1774-80. [Crossref] [PubMed]
- Eguchi T, Takahashi Y, Yamagata M, et al. Gastric cancer in young patients. J Am Coll Surg 1999;188:22-6. [Crossref] [PubMed]
- Buffart TE, Carvalho B, Hopmans E, et al. Gastric cancers in young and elderly patients show different genomic profiles. J Pathol 2007;211:45-51. [Crossref] [PubMed]
- Morgensztern D, Campo MJ, Dahlberg SE, et al. Molecularly targeted therapies in non-small-cell lung cancer annual update 2014. J Thorac Oncol 2015;10:S1-63. [Crossref] [PubMed]
- Wen M, Wang X, Sun Y, et al. Detection of EML4-ALK fusion gene and features associated with EGFR mutations in Chinese patients with non-small-cell lung cancer. Onco Targets Ther 2016;9:1989-95. [Crossref] [PubMed]
- Shi Y, Li J, Zhang S, et al. Molecular Epidemiology of EGFR Mutations in Asian Patients with Advanced Non-Small-Cell Lung Cancer of Adenocarcinoma Histology - Mainland China Subset Analysis of the PIONEER study. PLoS One 2015;10:e0143515. [Crossref] [PubMed]
- Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573-82. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [Crossref] [PubMed]
- Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015;36:E2423-9. [Crossref] [PubMed]
- Abbott KL, Nyre ET, Abrahante J, et al. The Candidate Cancer Gene Database: a database of cancer driver genes from forward genetic screens in mice. Nucleic Acids Res 2015;43:D844-8. [Crossref] [PubMed]


