
ZIC5 promotes human hepatocellular carcinoma cell proliferation through upregulating COL1A1
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor worldwide and the third leading cause of tumor-related deaths (1,2). Approximately 700,000 new cases of HCC are diagnosed every year, and more than half of them are Chinese patients (3,4). Currently, chronic hepatitis B virus infection is still considered as the main cause of HCC. In addition, chronic hepatitis C virus infection, eating food containing aflatoxin, alcoholism, and smoking are also important factors. In recent years, the incidence of diabetes and obesity has gradually increased, which are also high-risk factors for HCC that cannot be ignored (5,6). With the improvement of economic level and health awareness, as well as the expansion of the scope and intensity of hepatitis B screening, some HCC patients can be cured due to early detection and early treatment. However, due to the lack of obvious clinical manifestations in the early and middle stages of HCC, many patients are in the advanced stage when they are first diagnosed, losing the best time for treatment and contributing to poor prognosis. Therefore, elucidating the mechanism of the occurrence and development of HCC and developing clinical drugs for HCC have important clinical significance for improving the prognosis of advanced-stage HCC patients.
Zinc finger of the cerebellum (ZIC) family genes were first discovered in the cerebellum of adult mice and named after their abundant zinc finger protein expression (7). The ZIC gene family contains 5 members, namely ZIC1-5 genes (8). Prior studies have pointed out that ZIC genes are closely related to the occurrence and development of various tumors such as nervous system tumors, small cell lung cancer, endometrial tumors, gastrointestinal cancer, and liver cancer (9-12). ZIC5, as a member of the ZIC gene family, encodes a member of the C2H2-type zinc finger proteins which act as transcriptional repressors. ZIC5 is highly expressed in various types of tumors, resulting in rapid tumor progression and poor prognosis (13-19).
In this study, by analyzing ZIC5 expression in 33 types of tumors from The Cancer Genome Atlas (TCGA) database, we determined that ZIC5 is significantly upregulated in tumor tissues compared to normal tissues, including HCC. High ZIC5 expression in HCC was positively correlated with MKI67 and indicated poor prognosis. Our findings revealed that ZIC5 overexpression promoted the proliferation of HCC cells in vitro and significantly increased collagen type I alpha 1 (COL1A1) expression. We present the following article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-335/rc).
Methods
Bioinformatics analysis
ZIC5 mRNA expression levels and the correlation between ZIC5 and MKI67 in pan-cancer (including 33 types of cancers) were analyzed by UCSCXenaShiny, an R package for interactive analysis of UCSC Xena Data. Kaplan-Meier survival curves for the effect of ZIC5 mRNA expression level on HCC patient survival and COL1A1 mRNA levels in human HCC tissues (n=369) and normal liver tissues (n=160) were both obtained from Gene Expression Profiling Analysis (GEPIA) based on TCGA dataset (https://tcga-data.nci.nih.gov/tcga/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
The human HCC cell lines Huh1 and HepG2 were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Huh1 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; BasalMedia, Shanghai, China) and HepG2 cells were cultured in Minimum Essential Medium (MEM; BasalMedia, Shanghai, China) with 10% (v/v) fetal bovine serum (FBS; Biological Industries, Israel), 100 U/mL penicillin G, and 100 µg/mL streptomycin at 37 ℃ under a humidified atmosphere of 5% CO2.
Overexpression of ZIC5
The complete coding sequence (CDS) of the human ZIC5 gene was amplified and inserted into the coding region of the pcDNA3.1-3xflag vector (YouBio, Changsha, China). ZIC5 overexpression was prepared by transient transfection of the plasmid via Lipofectamine 2000 (Invitrogen, Waltham, MA, USA).
Cell proliferation assay
Cell proliferation was determined using cell counting kit-8 (CCK-8) assays (Dojindo, Kumamoto, Japan). Huh1 and HepG2 cells were seeded in 6-well plates at a density of 3.0×105 cells per well with 2 mL medium. Then, the cells were transiently transfected with the ZIC5 plasmid by Lipofectamine 2000. After 6–8 hours, the cells were reseeded in 96-well plates at a density of 4,000 cells per well. Subsequently, 10% (v/v) CCK-8 solution was added to the plates at 0, 24, 48, 72 hours, followed by another 2 hours incubation at 37 ℃. The absorbance was measured by a microplate reader (BioTek, Vermont, USA) at 450 nm. GraphPad Prism 5 was used to plot the growth curves.
Colony formation assays
Colony formation assays were performed to investigate the effect of ZIC5 on cell proliferation. Huh1 and HepG2 cells were seeded in 6-well plates at a density of 500 cells per well with 2 mL medium after transient transfection for 6–8 hours, and cultured for another 10 days with continuous replacement of fresh medium. Then, cells in 6-well plates were fixed in 4% paraformaldehyde (Solarbio, Beijing, China) for 20 minutes and stained with 1% crystal violet staining solution for 30 minutes. Cell colonies with more than 50 cells were counted and scanned.
RNA sequencing and analysis
Total RNA was extracted from Huh1 cells after transient transfection for 48 hours using a Spin Column Animal Total RNA Purification kit (Sangon Biotech, Shanghai, China). RNA concentrations were detected by a Nano-Photometer spectrophotometer (Implen, CA, USA). RNA was sequenced on an Illumina Novaseq platform (Novogene, Beijing, China). Given that there were no biological replicates, the edgeR program package was used to adjust the read counts through a scaling normalized factor before differential gene expression analysis. Then, differential gene expression analysis between the two conditions of Huh1 cells were executed by the edgeR R package (3.22.5). P values were adjusted using the Benjamini and Hochberg method. Fold change (FC) ≥2 and adjusted P value ≤0.05 were set as the thresholds for significantly differentially expressed genes (DEGs).
Real time quantitative reverse transcription polymerase chain reaction (real time qRT-PCR)
Total RNA was extracted using a Spin Column Animal Total RNA Purification kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. A total of 1 µg of RNA was used for reverse transcription by HiScript III RT SuperMix for qPCR (Vazyme, Nanjing, China). Real time PCR was conducted with a SYBR® Premix Ex Taq™ II kit (TaKaRa, Kusatsu, Japan). All procedures were performed according to the manufacturer’s instructions. Relative gene expression levels with the β-actin gene as an internal control were determined by the 2−ΔΔCt method (20). The primers used in the study were as follows: β-actin forward: TGGCACCCAGCACAATGAA, β-actin reverse: CTAAGTCATAGTCCGCCTAGAAGCA, ZIC5 forward: ACTGCAAGATTCGAGGCTGT, ZIC5 reverse: TGAGTAACCAAGGGGTCCTG; COL1A1 forward: GTGCTAAAGGTGCCAATGGT, COL1A1 reverse: ACCAGGTTCACCGCTGTTAC.
Western blot
Huh1 and HepG2 cells were collected by centrifugation, washed with PBS, lysed in RIPA buffer (Beyotime Biotechnology, Shanghai, China), and quantified by a BCA kit (Beyotime Biotechnology, Shanghai, China). Equal amounts of protein were subjected to 4–12% gradient polyacrylamide gel electrophoresis (PAGE; GenScript, Nanjing, China) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corp., Bedford, MA, USA). Subsequently, the membranes were blocked with 5% skimmed milk (Oxoid, UK) for 2 hours at room temperature and immunoblotted with the primary antibodies at 4 ℃ overnight. Finally, the immunoreactive bands were developed using an ECL detection system (BLT, PHOTO Technology, China). Gray-scale scanning was carried out by ImageJ software for relative quantitative analysis of proteins. The primary antibodies used in the study were as follows: β-actin (#AB0035, Abways), ZIC5 (#NBP2-84346, Novus), COL1A1 (#67288-1-Ig, Proteintech), vimentin (#CY5134, Abways), N-cadherin (#CY5015, Abways), E-cadherin (#CY1155, Abways), p21 (#2947, Cell Signaling Technology), Bcl2 (#CY6717, Abways), c-Myc (#CY5150, Abways).
Statistical analysis
Statistical analyses were conducted using Student’s t-test. Each experiment was repeated 3 times, and the quantitative data were presented as mean ± SEM. P values less than 0.05 were considered statistically significant.
Results
High ZIC5 expression was associated with poor prognosis of HCC
To further understand the role of ZIC5 in cancers, we analyzed its mRNA expression from pan-cancer datasets compiled using UCSC Xena. The analysis results showed that ZIC5 was abnormally expressed in a variety of cancers. ZIC5 expression had no significant difference in cancer vs. normal tissues only in a few cancers, including glioblastoma multiforme (GBM), kidney renal papillary cell carcinoma (KIRP), pheochromocytoma and paraganglioma (PCPG), sarcoma (SARC), skin cutaneous melanoma (SKCM), and thymoma (THYM) (Figure 1A). To emphasize the correlation between ZIC5 expression and cell proliferation, the relationship between the mRNA levels of ZIC5 and MKI67 were analyzed in the pan-cancer database. Pearson’s correlation analysis showed that the mRNA levels of ZIC5 were positively correlated with the mRNA levels of MKI67 in HCC, with r=0.57 and P<0.001 (Figure 1B, Table 1). Survival analysis of 90 cases with high ZIC5 expression and 275 cases with low/medium ZIC5 expression from TCGA dataset demonstrated that the group with high ZIC5 expression had a shorter overall survival time than the low/medium ZIC5 expression group, with P=0.00058 (Figure 1C). These data reveal the important clinical value of ZIC5 in HCC.

Table 1
| Type | Correlation coefficient | P value |
|---|---|---|
| LUSC | 0.58 | 7.84×10−50 |
| LIHC | 0.57 | 1.87×10−37 |
| HNSC | 0.46 | 1.03×10−30 |
| BLCA | 0.39 | 1.20×10−16 |
| PRAD | 0.34 | 1.21×10−16 |
| KIRC | 0.31 | 4.77×10−15 |
| LUAD | 0.31 | 1.79×10−14 |
| KIRP | 0.38 | 1.42×10−12 |
| STAD | 0.31 | 9.13×10−12 |
| CESC | 0.34 | 5.30×10−10 |
| BRCA | 0.17 | 2.48×10−9 |
| COAD | 0.31 | 6.01×10−9 |
| ESCA | 0.36 | 2.00×10−7 |
| CHOL | 0.58 | 2.50×10−5 |
| PAAD | 0.29 | 5.63×10−5 |
| TGCT | 0.33 | 7.99×10−5 |
| OV | 0.17 | 0.00 |
| ACC | 0.36 | 0.00 |
| UCEC | 0.23 | 0.00 |
| LGG | −0.14 | 0.00 |
| SARC | 0.17 | 0.00 |
| MESO | 0.29 | 0.00 |
| DLBC | 0.34 | 0.02 |
| READ | 0.21 | 0.03 |
| UVM | 0.20 | 0.08 |
| GBM | −0.13 | 0.09 |
| SKCM | 0.07 | 0.13 |
| PCPG | 0.08 | 0.26 |
| UCS | 0.13 | 0.33 |
| THYM | −0.06 | 0.48 |
| KICH | 0.05 | 0.64 |
| THCA | −0.01 | 0.73 |
| LAML | −0.00 | 0.99 |
ZIC5, Zinc finger of the cerebellum 5; LUSC, lung squamous cell carcinoma; LIHC, liver hepatocellular carcinoma; HNSC, head and neck squamous cell carcinoma; BLCA, bladder urothelial carcinoma; PRAD, prostate adenocarcinoma; KIRC, kidney renal clear cell carcinoma; LUAD, lung adenocarcinoma; KIRP, kidney renal papillary cell carcinoma; STAD, stomach adenocarcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; CHOL, cholangiocarcinoma; PAAD, pancreatic adenocarcinoma; TGCT, testicular germ cell tumors; OV, ovarian serous cystadenocarcinoma; ACC, adrenocortical carcinoma; UCEC, uterine corpus endometrial carcinoma; LGG, brain lower grade glioma; SARC, sarcoma; MESO, mesothelioma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; READ, rectum adenocarcinoma; UVM, uveal melanoma; GBM, glioblastoma multiforme; SKCM, skin cutaneous melanoma; PCPG, pheochromocytoma and paraganglioma; UCS, uterine carcinosarcoma; THYM, thymoma; KICH, kidney chromophobe; THCA, thyroid carcinoma; LAML, acute myeloid leukemia.
Upregulation of ZIC5 promoted the proliferation of HCC cells
To select cell lines with relatively low expression of ZIC5 for overexpression, the expression levels of ZIC5 in three HCC cell lines including Huh1, HepG2, and LM3 cells were detected by real time PCR and western blot. As illustrated in Figure 2A,2B, ZIC5 expression in LM3 cells was relatively higher than that in Huh1 and HepG2 cells. Therefore, ZIC5 was overexpressed in Huh1 and HepG2 cells by transient transfection of the plasmid. ZIC5 was successfully overexpressed in Huh1 and HepG2 cells (Figure 2C,2D). To explore the role of ZIC5 in the proliferation of HCC cells, CCK-8 assays and the colony formation assay were performed in vector and ZIC5 overexpressing cells.
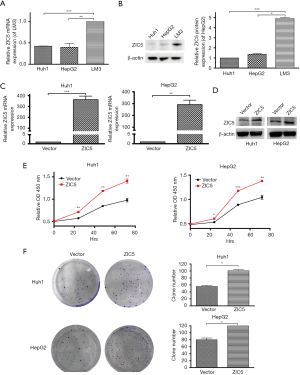
The proliferation of Huh1 and HepG2 cells was markedly increased by ZIC5 overexpression, especially at 48 and 72 hours (Figure 2E). Indeed, the colony formation ability of Huh1 and HepG2 cells was also increased after ZIC5 upregulation (Figure 2F). These findings suggest that ZIC5 upregulation can promote the proliferation of HCC cells.
ZIC5 promoted HCC cell proliferation through upregulating COL1A1
To explore the possible downstream target genes related to ZIC5 in the regulation of HCC cell proliferation, RNA sequencing was conducted in Huh1 cells with vector and ZIC5 overexpression. According to the screening conditions of DEGs described in the methods (FC ≥2 and adjusted P value ≤0.05), a total of 1,025 DEGs were obtained in Huh1 cells after ZIC5 overexpression, including 861 upregulated genes and 164 downregulated genes (Figure 3A). Among these DEGs, a regulator of hepatic fibrosis, COL1A1, which was significantly increased by ZIC5 overexpression in Huh1 cells, attracted our attention. COL1A1, as the major component of type I collagen, has been reported to be a potential prognostic biomarker for multiple malignant tumors. A series of recent studies indicated that downregulation of COL1A1 could inhibit the proliferation, migration, invasion, and promote the apoptosis of HCC cells (21-23). Moreover, the mRNA expression levels of COL1A1 were increased in Huh1 and HepG2 cells with ZIC5 overexpression as detected by real time PCR (Figure 3B). Then, we analyzed relative COL1A1 mRNA levels in TCGA LIHC dataset including 369 HCC tissues and 160 normal tissues and found that COL1A1 mRNA was significantly higher in HCC tissues than in normal tissues (Figure 3C).
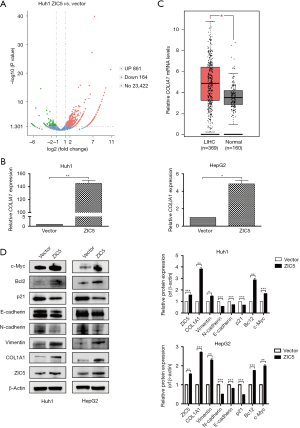
To investigate the effect of ZIC5 overexpression on the proliferation, migration, invasion, apoptosis, and cell cycle of HCC cells, the protein levels of related proteins were detected by western blot. Upon ZIC5 overexpression in Huh1 and HepG2 cells, we observed that the protein levels of COL1A1 were increased, consistent with its mRNA levels. In addition, the expression of c-Myc, a protein related to cell proliferation, was also increased. Increased expression of the anti-apoptotic protein Bcl2 could inhibit cell apoptosis. The decreased expression of p21, a cell cycle inhibitory protein, indicated the release of cell cycle inhibition. E-cadherin, N-cadherin, and vimentin are markers closely related to cell migration and invasion. ZIC5 overexpression suppressed the expression of E-cadherin and N-cadherin, while vimentin expression was increased (Figure 3D). The findings revealed that ZIC5 upregulation could accelerate the migration and invasion of HCC cells. In summary, ZIC5 may promote HCC cell proliferation by increasing COL1A1 expression.
Discussion
ZIC5, as a member of the ZIC gene family, has been reported to be associated with various tumors. In lung cancer, ZIC5 expression in tumors was much higher than that in adjacent tissues, and was positively correlated with differentiation and tumor size. Moreover, patients with high ZIC5 expression had shorter overall survival times (24). Notably, knockdown of ZIC5 inhibited non-small cell lung cancer cell proliferation and led to cell cycle arrest in G2 phase (16). Another study confirmed that ZIC5 expression was significantly increased in Chinese patients with colorectal cancer, and ZIC5 increased the proportion of cells in S phase to accelerate colorectal cancer cell proliferation (25). Knockdown of ZIC5 in colorectal cancer cells increased glycolytic status and tolerance to low-glucose conditions (26). As previously reported in the literature, ZIC5, as a transcriptional suppressor of CDH1, promoted melanoma cell proliferation, drug resistance, and in vivo metastasis by activating PDGFD and FAK (18). CircRNA hsa_circ_0007534 serves as an oncogene by sponging miR-761 to increase the expression of ZIC5, thus promoting glioma cell proliferation and migration (27). A study on the function of ZIC5 in HCC revealed that ZIC5 expression in HCC tissues was higher than that in normal tissues, and HCC patients with high ZIC5 expression had a poor prognosis (28). ZIC5 overexpression could promote the proliferation, migration, and invasion of HCC cells by increasing the expression of genes related to the Wnt/β-catenin signaling pathway and facilitating β-catenin entry into the nucleus (28). A series of recent studies indicated that ZIC5 may act as an oncogene in tumors (16,27,28). Although ZIC5 has been reported to be associated with various tumors, its role in HCC has only been reported in one study so far (28). Our study further explored the mechanism of ZIC5 in the occurrence and development of HCC. To elucidate the relationship between ZIC5 and various tumors, we analyzed its expression in various tumors from TCGA database. Our findings confirmed that ZIC5 was highly expressed in a variety of tumors, including HCC, while its expression had no significant difference in tumor tissues vs. normal tissues in only a few tumors. A series of recent studies indicated that high ZIC5 expression can promote cancer cell proliferation (16,18,27,28). The protein encoded by MKI67 is an antigen related to cell proliferation and is indispensable for cell proliferation. MKI67 is used as an indicator to detect cell proliferation. Therefore, we analyzed the relationship between ZIC5 and MKI67 expression in various tumors by Pearson’s correlation analysis. The results showed that ZIC5 mRNA levels were positively correlated with MKI67 mRNA levels in HCC tissues. Furthermore, we discovered that HCC patients with higher ZIC5 expression had a shorter survival time (Figure 1). Based on the above bioinformatics analysis, we selected HCC cells to validate these findings. By detecting the mRNA and protein levels of ZIC5 in Huh1, HepG2, and LM3 cells, we selected Huh1 and HepG2 cells with relatively low ZIC5 expression for further experiments. CCK-8 and colony formation assays determined that ZIC5 overexpression could promote HCC cell proliferation (Figure 2).
To further explore the underlying mechanism of ZIC5 in cell proliferation, we performed RNA sequencing of Huh1 cells overexpressing ZIC5. Analysis of the RNA sequencing results revealed that the expression of COL1A1, closely related to the occurrence and development of HCC, was increased after ZIC5 overexpression. The extracellular matrix (ECM) is a complex network structure of macromolecular substances that provides support and connections between tissues and cells (29). As an important part of the tumor microenvironment, the ECM plays critical roles in the development and metastasis of tumors (30,31). Collagen, as the major structural protein of the ECM, is involved in the formation of microfibrillar and fibrillar networks of basement membranes and ECM (32). COL1A1, as a common type of collagen, has been reported to be involved in the proliferation, migration, invasion, and prognosis of various tumors, such as HCC, breast cancer, ovarian cancer, astrocytoma, lung cancer, gastric cancer, mesothelioma, and colorectal cancer (21,33-38). In the present study, we found that upregulating ZIC5 levels could increase the expression of COL1A1, and the expression of COL1A1 in HCC tissues was significantly higher than that in normal tissues. Western blot analysis revealed that COL1A1 upregulation altered the expression levels of proteins related to proliferation, migration, invasion, cell cycle, and apoptosis, including decreased expression of E-cadherin, N-cadherin, and p21, and increased expression of c-Myc, vimentin, Bcl2, and p21 (Figure 3).
In summary, our study reveals a novel mechanism of ZIC5 in promoting HCC cell proliferation. ZIC5 promotes HCC cell proliferation, migration, and invasion by increasing COL1A1 expression. This indicates that ZIC5 may be a potential therapeutic target for HCC. Although we have not conducted in-depth research on its underlying mechanism, the current findings suggest a correlation between ZIC5 and COL1A1, which provides a new perspective for follow-up studies.
Conclusions
Our research confirmed that, compared with adjacent normal tissues, the expression of ZIC5 was significantly higher in HCC tissues. Patients with high ZIC5 expression had a shorter overall survival time. ZIC5 overexpression promoted the proliferation, migration, and invasion of HCC cells by upregulating COL1A1. These results reveal the role of ZIC5 in HCC and propose a relationship between ZIC5 and COL1A1 expression for the first time.
Acknowledgments
Funding: The research was supported by the National Natural Science Foundation of China (No. 82104204), the Climbing Foundation of National Cancer Center (No. NCC201816B047), the Science and Technology Research Project of Henan Province, China (Nos. 212102311036 and 212102310325), and the Medical Science and Technology Research Project of Henan Province, China (No. LHGJ20200169).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-335/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-335/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-335/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Rinaldi L, Vetrano E, Rinaldi B, et al. HCC and Molecular Targeting Therapies: Back to the Future. Biomedicines 2021;9:1345. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Wang J, Li J, Tang G, et al. Clinical outcomes and influencing factors of PD-1/PD-L1 in hepatocellular carcinoma. Oncol Lett 2021;21:279. [Crossref] [PubMed]
- Alqahtani A, Khan Z, Alloghbi A, et al. Hepatocellular Carcinoma: Molecular Mechanisms and Targeted Therapies. Medicina (Kaunas) 2019;55:526. [Crossref] [PubMed]
- Shibata T. Genomic landscape of hepatocarcinogenesis. J Hum Genet 2021;66:845-51. [Crossref] [PubMed]
- Aruga J, Yokota N, Hashimoto M, et al. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J Neurochem 1994;63:1880-90. [Crossref] [PubMed]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci 2004;26:205-21. [Crossref] [PubMed]
- Houtmeyers R, Souopgui J, Tejpar S. Deregulation of ZIC Family Members in Oncogenesis. Adv Exp Med Biol 2018;1046:329-38. [Crossref] [PubMed]
- Hatayama M, Aruga J. Role of Zic Family Proteins in Transcriptional Regulation and Chromatin Remodeling. Adv Exp Med Biol 2018;1046:353-80. [Crossref] [PubMed]
- Lv Z, Qi L, Hu X, et al. Zic Family Member 2 (ZIC2): a Potential Diagnostic and Prognostic Biomarker for Pan-Cancer. Front Mol Biosci 2021;8:631067. [Crossref] [PubMed]
- Han Z, Jia J, Lv Y, et al. Transcriptional expression of ZICs as an independent indicator of survival in gliomas. Sci Rep 2021;11:17532. [Crossref] [PubMed]
- Satow R, Inagaki S, Kato C, et al. Identification of zinc finger protein of the cerebellum 5 as a survival factor of prostate and colorectal cancer cells. Cancer Sci 2017;108:2405-12. [Crossref] [PubMed]
- Han W, Zhang C, Gao XJ, et al. Clinicopathologic and Prognostic Significance of the Zinc Finger of the Cerebellum Family in Invasive Breast Cancer. J Breast Cancer 2018;21:51-61. [Crossref] [PubMed]
- Nyholm MK, Wu SF, Dorsky RI, et al. The zebrafish zic2a-zic5 gene pair acts downstream of canonical Wnt signaling to control cell proliferation in the developing tectum. Development 2007;134:735-46. [Crossref] [PubMed]
- Sun Q, Shi R, Wang X, et al. Overexpression of ZIC5 promotes proliferation in non-small cell lung cancer. Biochem Biophys Res Commun 2016;479:502-9. [Crossref] [PubMed]
- Zhang Y, Xi XT, Zhao X. Effect of zic5 on biological behaviors of prostate cancer and its mechanisms. Journal of Tongji University (Medical Science) 2018;39:61-5.
- Satow R, Nakamura T, Kato C, et al. ZIC5 Drives Melanoma Aggressiveness by PDGFD-Mediated Activation of FAK and STAT3. Cancer Res 2017;77:366-77. [Crossref] [PubMed]
- Sun J, Yoon J, Lee M, et al. Zic5 stabilizes Gli3 via a non-transcriptional mechanism during retinal development. Cell Rep 2022;38:110312. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Ma HP, Chang HL, Bamodu OA, et al. Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis. Cancers (Basel) 2019;11:786. [Crossref] [PubMed]
- Zhao W, Jiang X, Yang S. lncRNA TUG1 Promotes Cell Proliferation, Migration, and Invasion in Hepatocellular Carcinoma via Regulating miR-29c-3p/COL1A1 Axis. Cancer Manag Res 2020;12:6837-47. [Crossref] [PubMed]
- Hayashi M, Nomoto S, Hishida M, et al. Identification of the collagen type 1 α 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC Cancer 2014;14:108. [Crossref] [PubMed]
- Dong C, Li X, Li K, et al. The Expression Pattern of ZIC5 and its Prognostic Value in Lung Cancer. Cancer Biother Radiopharm 2021;36:407-11. [Crossref] [PubMed]
- Maimaiti A, Aizezi A, Anniwaer J, et al. Zinc finger of the cerebellum 5 promotes colorectal cancer cell proliferation and cell cycle progression through enhanced CDK1/CDC25c signaling. Arch Med Sci 2021;17:449-61. [Crossref] [PubMed]
- Zhao Z, Wang L, Bartom E, et al. β-Catenin/Tcf7l2-dependent transcriptional regulation of GLUT1 gene expression by Zic family proteins in colon cancer. Sci Adv 2019;5:eaax0698. [Crossref] [PubMed]
- Li GF, Li L, Yao ZQ, et al. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem Biophys Res Commun 2018;499:765-71. [Crossref] [PubMed]
- Liu L, Hu X, Sun D, et al. ZIC5 facilitates the growth of hepatocellular carcinoma through activating Wnt/β-catenin pathway. Biochem Biophys Res Commun 2018;503:2173-9. [Crossref] [PubMed]
- Gupta R. Epigenetic regulation and targeting of ECM for cancer therapy. Am J Physiol Cell Physiol 2022;322:C762-8. [Crossref] [PubMed]
- Mohan V, Das A, Sagi I. Emerging roles of ECM remodeling processes in cancer. Semin Cancer Biol 2020;62:192-200. [Crossref] [PubMed]
- Joshi RS, Kanugula SS, Sudhir S, et al. The Role of Cancer-Associated Fibroblasts in Tumor Progression. Cancers (Basel) 2021;13:1399. [Crossref] [PubMed]
- Huang J, Zhang L, Wan D, et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct Target Ther 2021;6:153. [Crossref] [PubMed]
- An Q, Liu T, Wang MY, et al. circKRT7-miR-29a-3p-COL1A1 Axis Promotes Ovarian Cancer Cell Progression. Onco Targets Ther 2020;13:8963-76. [Crossref] [PubMed]
- Liu J, Shen JX, Wu HT, et al. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med 2018;25:211-23. [PubMed]
- Zhang Z, Wang Y, Zhang J, et al. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol Med Rep 2018;17:5037-42. [Crossref] [PubMed]
- Sun S, Wang Y, Wu Y, et al. Identification of COL1A1 as an invasion-related gene in malignant astrocytoma. Int J Oncol 2018;53:2542-54. [Crossref] [PubMed]
- Geng Q, Shen Z, Li L, et al. COL1A1 is a prognostic biomarker and correlated with immune infiltrates in lung cancer. PeerJ 2021;9:e11145. [Crossref] [PubMed]
- Zhang C, Liu S, Wang X, et al. COL1A1 Is a Potential Prognostic Biomarker and Correlated with Immune Infiltration in Mesothelioma. Biomed Res Int 2021;2021:5320941. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


