
Proteogenomic analysis in an early onset diffuse gastric cancer patient revealed alterations in PIK3R1, TP53, SMAD4 and a potential role of the PI3K-AKT and EGFR pathways: a case report
Introduction
Typically, early-onset gastric cancers (EOGC) present at the age of 45 years old or younger. Although its prevalence ranges from 2.7% to 15% depending on the population being studied, it is generally accepted that 10% of gastric cancers (GCs) fall into this category. However, using the SEER database and a <60 years old cutoff, a recent study determined that >30% of GC cases in the United States were EOGC during the period 1973–2015. This study also found that EOGC was associated with the presence of signet-ring cells, diffuse histology and metastatic disease, relative to “late onset” cases (1). Intriguingly, despite its early clinical onset that would suggest a strong hereditary component, reports show that only a small proportion of cases (~10%) are associated with germinal mutations.
To date, cancer heterogeneity remains a major obstacle for the development of effective therapies. In recent decades, the development of massively-parallel sequencing techniques [such as next generation sequencing (NGS) and RNA-sequencing (RNASeq)] has allowed the identification of distinctive molecular signatures in several malignancies, including GC; thereby establishing molecular subtypes of the disease. In this regard, one of the best examples is the classification elaborated by The Cancer Genome Atlas Research Network Group (TCGA) (2). Molecular characterization studies in EOGC patients have shown high rates of CDH1 but similar rates of TP53 mutations (3) compared to “traditional” GC, along with a higher frequency of Epstein-Barr virus positive (EBV+) and genomically stable (GS) molecular subtypes (1). Moreover, a related study found a lower frequency of RHOA somatic mutations in EOGC (4). Interestingly, unlike “traditional” GC studies have also demonstrated higher mutation rates on MUC5B and BANP (5).
Herein, we report clinical and proteogenomic characteristics of a 26-year-old EOGC patient with a tubular poorly cohesive cell adenocarcinoma characterized by rapid clinical deterioration and unfavorable prognosis. We present the following case in accordance with the CARE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-780/rc).
Case presentation
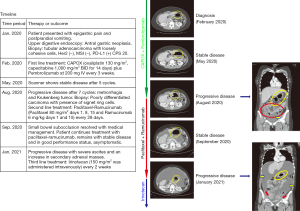
The patient was a 26-year-old female with a history of insulin resistance and a laparoscopic cholecystectomy for gallstones. In January 2020 patient came to the clinic after 3-month of episodic epigastric pain and post-prandial vomiting. Upper digestive endoscopy revealed an antral gastric neoplasia. Endoscopic biopsy confirmed a tubular adenocarcinoma with poorly cohesive cells. Molecular studies demonstrated HER2 negativity, MMR proficiency and PD-L1/combined positive score (CPS) =20. Timeline of relevant therapies and clinical follow-up is summarized in Figure 1 (left panel). Computed tomography (CT) scan showed thickening of the gastric mucosa, epigastric and infra-pyloric adenopathies and peritoneal carcinomatosis (Figure 1, right panels); in February 2020, patient started first-line CAPOX + pembrolizumab in the context of a clinical trial. After 5 cycles, CT scan showed stable disease. At cycle 7, the patient developed metrorrhagia and abdominal pain. In August 2020 a gynecological ultrasound discovered bilateral adnexal masses. Scanner confirmed progression of peritoneal carcinomatosis. Biopsy revealed a signet ring cell+/poorly differentiated carcinoma. The same biopsy sample was used for a proteogenomic study. Then in August 2020, patient started second-line paclitaxel plus ramucirumab for metastatic gastric cancer. A week after the patient was hospitalized for partial intestinal occlusion in the small bowel eventually resolved by systemic corticosteroids. The patient displayed good tolerance to paclitaxel plus ramucirumab and maintained performance status without symptoms until January 2021 when a follow-up CT scan showed an increase in ascites and secondary adnexal masses. Following progression, clinical trial blind information was open to decide next treatment (third line). Patient then received palliative irinotecan and remained under this treatment until April 14th, 2021, when she passed away.
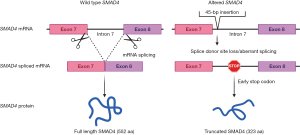
As mentioned, a NGS panel of 688 cancer-related genes (Sentis Cancer + Discovery, BGI) found 18 somatic mutations in a GC biopsy (FFPE) of the patient. Three of them were clinically relevant and 15 were variants of uncertain significance (VUS). One germinal likely pathogenic mutation was found. Tumor mutational burden (TMB) was 4.3 mut/MB and classified as microsatellite stable (MSS). Relevant alterations included a TP53 mutation, p.H233_Y234insSIH, a 3-aminoacid insertion in the DNA binding domain of p53, classified as likely pathogenic and a p53 copy number loss (Table 1). Analyses also found a PIK3R1 nonsense, likely pathogenic mutation, (p.Q110*) and a germline previously unreported intronic mutation in the SMAD4 gene. This alteration is caused by a 45-nucleotide insertion in the exon 7—intron 7 boundary that alters the wild type donor site, probably affecting splicing (Figure 2). Interestingly, skipping of intron 7 and the use of alternative splicing donor sites causes an early stop codon that translates into a truncated protein of 323 amino acids, versus 552 in the full-length protein. The truncated protein loses the MH2 domain in SMAD4, which is required for homomeric and heteromeric interactions and transcriptional regulation. Please note that our in-silico prediction of functional impact of this alteration should be further validated.
Table 1
| Affected gene | Type of alteration | Category | Approved therapies | Functional consequence |
|---|---|---|---|---|
| TP53 | Insertion | Somatic, likely pathogenic | None | Impaired protein and DNA binding; impaired p53 function |
| TP53 | Copy number loss | Somatic, likely pathogenic | None | TP53 deletion and poorer survival |
| PIK3R1 | Non-sense mutation | Somatic, likely pathogenic | None | Under-expression and poorer survival |
| SMAD4 | Intronic insertion | Germinal, likely pathogenic | None | Aberrant mRNA splicing leads to truncated form of protein |
NGS, next generation sequencing.
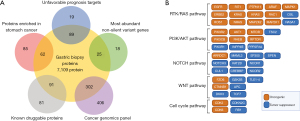
In addition, we performed an exploratory qualitative proteomic analysis seeking to identify relevant proteins in the biology and pharmacology of this aggressive type of GC. A workflow of the proteomic analysis can be found in Figure S1. Briefly, a fresh frozen biopsy from the same tumor was used for the proteomic profile characterization. A total of 7,109 proteins were identified, among these 25 have been reported as “most abundant non-silent variant genes”, 89 as “unfavorable prognosis targets”, and 91 as “known druggable proteins” (6). Also, 62 out of 147 were identified as “proteins enriched in stomach cancer” according to The Human Protein Atlas (7) (Figure 3A). Regarding potential druggable targets, we identified AKT1/mTOR and EGFR pathways; Figure 3B show oncogenic and tumor suppression proteins discovered using our approach and the signaling pathway they belong. Unfortunately, our approach did not allow us to identify under/over expressed pathways/proteins via a quantitative or specific phosphoproteomic analysis. A full list of identified proteins can be found in website (https://cdn.amegroups.cn/static/application/011747afd4dd140646bb20a12cac8bce/jgo-21-780-1.xlsx).
Ethical statement
All procedures performed in the study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
Early-onset GC incidence has steadily increased over the last 4 decades (1). A recent population-based cohort study assessed global GC incidence and mortality during the 1980–2018 period and reported an increase in GC incidence among <40 years old individuals in several countries including the United Kingdom, Sweden and Ecuador (8). Studies suggest that this increase is not linked to traditional risk factors that affect GC incidence such as binge drinking or smoking (9,10). Moreover, EOGC seems to display distinct clinico-pathological characteristics versus traditional GC. Studies report a predominance of the GS subtype in EOGC, which is characterized by poor prognosis, and a lack of therapeutic benefit by traditional systemic treatments such as chemotherapy (11). Current literature indicates that EOGC is more frequent in females (12-14), generally of diffuse type (14), multifocal (15) and non-microsatellite instability (MSI) (16,17). It is also estimated that about 10% of cases have a family history (9). Accordingly, the patient on this report was a female, MSS, HER2−, and therefore did not qualify for standard targeted therapies, like trastuzumab. As occurs with “traditional” or “ordinary” GC (18), ethnicity appears to be a significant risk factor for EOGC. A retrospective study that included >3,800 EOGC cases in Hispanic/Latino-origin US residents found that Mexican or Central/South American patients were more likely to be diagnosed with EOGC versus Cuban, Puerto Rican or Dominican Republic counterparts (19). In fact, the percentage of <40 years. Patients among Mexicans or Central/South Americans was 15% or 11%, respectively versus 3% in the entire cohort of the National Cancer Database, used as reference for this study.
Although our NGS analyses did not find relevant actionable targets, our proteomic analysis identified AKT/mTOR and EGFR as potential actionable pathways, which partially explains the rapid progression (1). Usually, EOGCs are linked to the EBV+ subtype and immunotherapy response, however this patient was categorized as EBV− and remained with stable disease for a relatively short period (5 cycles) after receiving CAPOX plus pembrolizumab. Similarly, a CPS =20 would also suggest immunotherapy response (20). Unfortunately, this was not the case. A report demonstrated a high spatial and temporal heterogeneity in PD-L1 expression in gastroesophageal adenocarcinoma (21). Thus, a potential explanation could be a rapid clonal expansion of tumor cells that became resistant to the checkpoint inhibitor.
Evidently, sporadic EOGC cases or those with a weak family history cannot be explained by traditional genetics. Our report is a case in point. Proteomic or epigenetic analyses could offer an alternative approach to discover potential therapies. Unfortunately, given the limitations of our proteomic analysis we were unable to test the clinical efficacy of targeting the AKT/mTOR or the EGFR pathways in this patient. Notably, a previous case report demonstrates long term survival in a metastatic GC patient by the combined use of cetuximab and FOLFIRI (22). On the other hand, in line with our findings, investigators have also speculated that PIK3CA or AKT inhibitors could be a promising alternative for GC patients (23,24). Obviously, more comprehensive studies should explore other alternatives such as DNA methylation in EOGC.
In general, it is assumed that younger patients are exposed to fewer environmental carcinogens and therefore the underlying causes of EOGC are mainly genetic, however as mentioned above only 10% of cases report a family history. Interestingly, the presence of a germline SMAD4 mutation in this patient suggests a hereditary component on EOGC. SMAD4 (formerly called DPC4) is widely recognized as a tumor suppressor gene, frequently mutated in pancreatic (25) and colon (26) cancers. In mice, heterozygous null SMAD4 mutants (SMAD4+/−) develop GI polyposis and tumors that overexpress Cyclin D1 and TGF-β (27). Furthermore, concomitant loss of CDH1 and SMAD4 in mice causes nuclear accumulation of β-catenin that promotes diffuse-type gastric adenocarcinomas and metastasis (28). In humans, a study reports 21% of juvenile polyposis syndrome (JPS) patients harbored germline SMAD4 alterations. Unlike our study, SMAD4 mutations were detected in exons 1, 4, 8, 10 and 11; investigators speculate that a wide range of SMAD4 alterations can affect TGF-β signaling (29). A similar study found a total of 17 germline SMAD4 mutations and six large deletions in a cohort of 80 JPS patients, among these 11 were predicted to generate a truncated protein (30). Moreover, a systematic analysis identified SMAD4 as a GC susceptibility gene (31). Besides germinal alterations, somatic SMAD4 mutations are common among poorly cohesive gastric carcinoma patients (32). Finally, a study that analyzed 80 sporadic early onset diffuse gastric cancer patients by whole exome sequencing found a high prevalence of CDH1, TP53, PIK3CA and TGFBR1 somatic mutations (4) partially confirming our findings. Interestingly, somatic CDH1 and TGFBR1 mutations were more frequently observed among females.
Conclusions
In summary, EOGC are hard-to-treat, poor prognosis malignancies with an increasing incidence. Tumor mutational profiling in this refractory EOGC patient did not identify potentially actionable relevant targets. In contrast, a proteomic profiling identified signaling pathways with therapeutic potential (AKT/mTOR and EGFR). Unfortunately, given the rapid deterioration of this patient the clinical utility of this approach could not be assessed. We present a model to search for new molecular alterations or targets in refractory patients. Future studies should clarify the benefits (if any) of a combined proteogenomic approach in the treatment refractory EOGC patients.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-780/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-21-780/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bergquist JR, Leiting JL, Habermann EB, et al. Early-onset gastric cancer is a distinct disease with worrisome trends and oncogenic features. Surgery 2019;166:547-55. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Setia N, Wang CX, Lager A, et al. Morphologic and molecular analysis of early-onset gastric cancer. Cancer 2020;127:103-14. [Crossref] [PubMed]
- Cho SY, Park JW, Liu Y, et al. Sporadic Early-Onset Diffuse Gastric Cancers Have High Frequency of Somatic CDH1 Alterations, but Low Frequency of Somatic RHOA Mutations Compared With Late-Onset Cancers. Gastroenterology 2017;153:536-49.e26. [Crossref] [PubMed]
- Mun DG, Bhin J, Kim S, et al. Proteogenomic Characterization of Human Early-Onset Gastric Cancer. Cancer Cell 2019;35:111-24.e10. [Crossref] [PubMed]
- Ge S, Xia X, Ding C, et al. A proteomic landscape of diffuse-type gastric cancer. Nat Commun 2018;9:1012. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Wong MCS, Huang J, Chan PSF, et al. Global Incidence and Mortality of Gastric Cancer, 1980-2018. JAMA Netw Open 2021;4:e2118457. [Crossref] [PubMed]
- Milne AN, Sitarz R, Carvalho R, et al. Early onset gastric cancer: on the road to unraveling gastric carcinogenesis. Curr Mol Med 2007;7:15-28. [Crossref] [PubMed]
- Ma Z, Liu X, Paul ME, et al. Comparative investigation of early-onset gastric cancer. Oncol Lett 2021;21:374. [Crossref] [PubMed]
- Sohn BH, Hwang JE, Jang HJ, et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin Cancer Res 2017;23:4441-9. [Crossref] [PubMed]
- Derakhshan MH, Liptrot S, Paul J, et al. Oesophageal and gastric intestinal-type adenocarcinomas show the same male predominance due to a 17 year delayed development in females. Gut 2009;58:16-23. [Crossref] [PubMed]
- Maeta M, Yamashiro H, Oka A, et al. Gastric cancer in the young, with special reference to 14 pregnancy-associated cases: analysis based on 2,325 consecutive cases of gastric cancer. J Surg Oncol 1995;58:191-5. [Crossref] [PubMed]
- Matley PJ, Dent DM, Madden MV, et al. Gastric carcinoma in young adults. Ann Surg 1988;208:593-96. [Crossref] [PubMed]
- Furukawa H, Iwanaga T, Imaoka S, et al. Multifocal gastric cancer in patients younger than 50 years of age. Eur Surg Res 1989;21:313-8. [Crossref] [PubMed]
- Lim S, Lee HS, Kim HS, et al. Alteration of E-cadherin-mediated adhesion protein is common, but microsatellite instability is uncommon in young age gastric cancers. Histopathology 2003;42:128-36. [Crossref] [PubMed]
- Carvalho R, Milne AN, van Rees BP, et al. Early-onset gastric carcinomas display molecular characteristics distinct from gastric carcinomas occurring at a later age. J Pathol 2004;204:75-83. [Crossref] [PubMed]
- Dong E, Duan L, Wu BU. Racial and Ethnic Minorities at Increased Risk for Gastric Cancer in a Regional US Population Study. Clin Gastroenterol Hepatol 2017;15:511-7. [Crossref] [PubMed]
- Karalis JD, Ju MR, Mansour JC, et al. The presentation of Hispanic gastric cancer patients varies by location of patient ancestry. J Surg Oncol 2021;124:1051-9. [Crossref] [PubMed]
- Wainberg ZA, Fuchs CS, Tabernero J, et al. Efficacy of Pembrolizumab Monotherapy for Advanced Gastric/Gastroesophageal Junction Cancer with Programmed Death Ligand 1 Combined Positive Score ≥10. Clin Cancer Res 2021;27:1923-31. [Crossref] [PubMed]
- Zhou KI, Peterson B, Serritella A, et al. Spatial and Temporal Heterogeneity of PD-L1 Expression and Tumor Mutational Burden in Gastroesophageal Adenocarcinoma at Baseline Diagnosis and after Chemotherapy. Clin Cancer Res 2020;26:6453-63. [Crossref] [PubMed]
- Adua D, Di Fabio F, Rojas Llimpe FL, et al. Long-term survival in an advanced gastric cancer patient treated with cetuximab in association with FOLFIRI: a case report. J Gastrointest Oncol 2014;5:E13-17. [PubMed]
- Yang J, Nie J, Ma X, et al. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer 2019;18:26. [Crossref] [PubMed]
- Singh SS, Yap WN, Arfuso F, et al. Targeting the PI3K/Akt signaling pathway in gastric carcinoma: A reality for personalized medicine? World J Gastroenterol 2015;21:12261-73. [Crossref] [PubMed]
- Xing S, Yang H, Liu J, et al. Prognostic Value of SMAD4 in Pancreatic Cancer: A Meta-Analysis. Transl Oncol 2016;9:1-7. [Crossref] [PubMed]
- Salovaara R, Roth S, Loukola A, et al. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut 2002;51:56-9. [Crossref] [PubMed]
- Xu X, Brodie SG, Yang X, et al. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene 2000;19:1868-74. [Crossref] [PubMed]
- Park JW, Jang SH, Park DM, et al. Cooperativity of E-cadherin and Smad4 loss to promote diffuse-type gastric adenocarcinoma and metastasis. Mol Cancer Res 2014;12:1088-99. [Crossref] [PubMed]
- Woodford-Richens K, Bevan S, Churchman M, et al. Analysis of genetic and phenotypic heterogeneity in juvenile polyposis. Gut 2000;46:656-60. [Crossref] [PubMed]
- Aretz S, Stienen D, Uhlhaas S, et al. High proportion of large genomic deletions and a genotype phenotype update in 80 unrelated families with juvenile polyposis syndrome. J Med Genet 2007;44:702-9. [Crossref] [PubMed]
- McKinley SK, Singh P, Yin K, et al. Disease spectrum of gastric cancer susceptibility genes. Med Oncol 2021;38:46. [Crossref] [PubMed]
- Kwon CH, Kim YK, Lee S, et al. Gastric poorly cohesive carcinoma: a correlative study of mutational signatures and prognostic significance based on histopathological subtypes. Histopathology 2018;72:556-68. [Crossref] [PubMed]


