
Expression characteristics of long non-coding RNA in colon adenocarcinoma and its potential value for judging the survival and prognosis of patients: bioinformatics analysis based on The Cancer Genome Atlas database
Introduction
Colon adenocarcinoma (COAD) is a primary malignant tumor in the colonic epithelium. The early symptoms of COAD are insignificant, and as it progresses, patients gradually present with symptoms such as abdominal pain, hematochezia, changes in stool characteristics, and body weight loss. At present, surgery-based comprehensive treatment options for colon cancer have the best efficacy, but for patients with advanced or end stage colon cancer, their therapeutic effect is still poor (1). Therefore, it is important to study the pathogenesis, early diagnostic markers, and molecular targets for targeted therapy of COAD. Long non-coding RNA (lncRNA) are RNA with a total nucleotide length of more than 200, and although they are not involved in gene transcription and play an important role in the development, mechanical homeostasis, and maintenance of cell fate, the abnormal expression of lncRNAs in different types of tumors and tissues is closely related to the development of cancer, so they provide new biomarkers and drug targets (2,3). At present, there have been an escalating number of studies on the relationship between specific lncRNA and the development of COAD. Dysregulation of lncRNA H19, SNHG7, HOTAIR, CRNDE are the lncRNAs which have been proved to affect the proliferation of cancer cells and predict bad prognosis (4-7). However, the down-regulation of lncRNA BDNF-AS in colorectal patients was seen as a protective gene which can inhibit tumor progression (8). With the development of whole exome sequencing, people can capture and enrich the DNA in the exon region of the whole genome, and then conduct high-throughput sequencing to find the genetic mutations related to protein function variation (9). Meanwhile, the abnormal expression of LncRNA gene profile in colon cancer can also reflect the changes related to protein function variation, but there is still a lack of comprehensive and macro investigation and analysis. With the development of next-generation sequencing (NGS) technology, RNA sequencing (RNA-seq) data has proliferated, and a large number of RNA-seq data can be downloaded from different public databases for analysis. The Cancer Genome Atlas (TCGA) database includes common cancer genome, transcriptome, proteome, and epigenetic group data and their associated clinical data, providing massive data for mining meaningful genomic changes and discovering biological mechanisms such as tumor development and progression (10,11). Downloading COAD-related RNA-seq data from TCGA for analysis of lncRNA expression characteristics in COAD tissues can preliminarily elucidate the mechanism of lncRNA in colorectal cancer (CRC) development and its potential application value in prognosis. There have been reports of mining biomarkers related to breast cancer, pancreatic cancer, and liver cancer using TCGA database, but data mining on COAD is rarely reported (12,13). Therefore, in this study, RNA-seq data from TCGA will be used to investigate lncRNA associated with COAD pathogenesis and to explore its application value in COAD diagnosis and prognosis. We present the following article in accordance with the REMARK reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-384/rc).
Methods
Data acquisition
We visited the Xena official website (https://xenabrowser.net/datapages/); RNAseq clinical expression profile data of COAD samples in TCGA database were obtained, with a total of 494 samples (including 453 tumor tissue samples and 41 normal tissue samples), each containing 60,489 different types of gene. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data processing and differential expression analysis
We downloaded RNA-seq expression profile data and extracted the raw counts expression matrix. Then, lncRNA genes (lincRNA, processed pseudogene, transcribed processed pseudogene, antisense RNA) were filtered from the data using R language (version 4.1.3; The R Foundation for Statistical Computing, Vienna, Austria). The matrix was divided into 2 groups “Tumor” and “Normal” according to the last 3 bits “01A” and “11A” of tissue listed in the matrix. Differential analysis was performed on the 2 groups of gene matrix using the “DESeq2” module of BiocManager software package in R; the screening threshold was |log2FC|>3, false discovery rate (FDR)<0.001, and eligible genes were considered significantly differentially expressed. Volcano plots were drawn using 2 modules: “ggpubr” and “ggthemes”.
Prognostic survival analysis
The survival data of 469 samples of colon cancer phenotype were obtained from the same Xena website, among which the expression profiles of lncRNA genes were selected. After combining the data with differentially expressed genes (DEGs), Cox regression analysis was performed using the “survival” package in R language, and the difference was considered significant at P<0.05 to determine the DEGs that had an effect on patient survival, and forest plots were drawn using “forestplot”. The median expression values of lncRNA in these patient samples divided the patients into a high expression group and low expression group. The survival rate differences of patients between high and low expression groups were compared. The Kaplan-Meier (KM) survival curve of lncRNA, which predicts the potential impact on the prognosis and survival time of patients, was drawn using the R survival package and the curve was subjected to logrank test. A P value <0.05 indicated that the expression level of lncRNA had a statistically significant difference in the prognosis and survival between the 2 groups.
LncRNA functional analysis
The prognosis-related genes were identified from survival analysis, and Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to screen the results with P<0.05 as significant enrichment results by coding genes containing the regulation of lncRNA using the “clusterprofiler” package in R.
Statistical analysis
All the differentially expressed data were analyzed by using R software (version 4.1.3) through the “DESeq2” and “survival” package. KM survival analysis (log-rank test) was applied. ROC curve analysis, univariate, and multivariate Cox regression analysis were conducted and P value <0.05 was considered as a significant threshold.
Results
Differential analysis expression and volcano plot display
According to the screening threshold, 204 significantly differentially lncRNA-expressed genes were obtained, among which, 156 were up-regulated and 48 were down-regulated. The volcano plot is shown in Figure 1, and the genes were ranked within 10 bits according to their FDR values, as shown in Table 1. We selected the 3 genes with the most significant expression levels, Linc02418, Blacat1, and Al353801.1, and obtained their expression levels in tumor tissues and normal tissues using punctate grouping comparison plots, as shown in Figure 2.
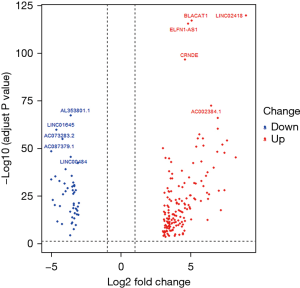
Table 1
| Gene | Ensembl ID | Log2 FC | FDR | Change |
|---|---|---|---|---|
| LINC02418 | ENSG00000214039.8 | 9.00 | 3.17e-120 | UP |
| BLACAT1 | ENSG00000281406.1 | 5.10 | 1.10e-117 | UP |
| ELFN1-AS1 | ENSG00000236081.1 | 4.85 | 4.93e-116 | UP |
| CRNDE | ENSG00000245694.9 | 4.61 | 4.16e-97 | UP |
| AC002384.1 | ENSG00000228742.9 | 6.49 | 5.32e-73 | UP |
| AL353801.1 | ENSG00000223462.2 | -3.61 | 5.84e-68 | DOWN |
| LINC01645 | ENSG00000224968.1 | -4.63 | 1.99e-60 | DOWN |
| AC073283.2 | ENSG00000226087.1 | -4.21 | 1.25e-55 | DOWN |
| AC087379.1 | ENSG00000254645.1 | -5.00 | 3.77e-49 | DOWN |
| LINC00484 | ENSG00000235641.4 | -3.63 | 5.20e-46 | DOWN |
lncRNA, long non-coding RNA; FC, fold change; FDR, false discovery rate.

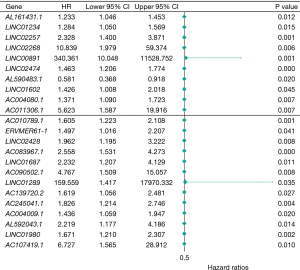
Cox analysis
Cox regression analysis was performed for the above 204 lncRNA genes using the “survival” package in R language, together with survival data. Using P<0.05 as the threshold, 23 lncRNA genes with significant effects on the prognosis and survival rate of patients were obtained, the forest plot is shown in Figure 3.
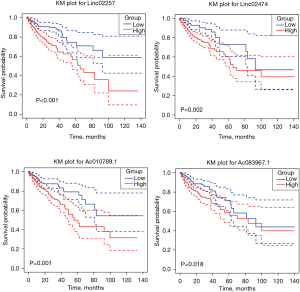
Survival and prognostic analysis
In all, 5 genes with P≤0.001 in Cox analysis, Linc02257, Linc00891, Linc02474, Ac010789.1, and Ac083967.1, were divided into high- and low-expression groups by median expression, and the KM curve was plotted using the survival package of R language. The KM curve of LINC00891 gene was not statistically significant (P=0.877), and the KM curve of the other 4 genes was statistically significant (P<0.05), as shown in Figure 4.
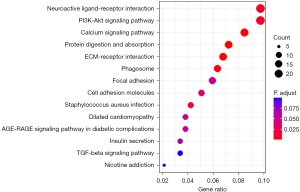
LncRNA enrichment functional analysis
The gene Linc02257 was selected for GO function and KEGG pathway enrichment analysis, which revealed that the inheritance of Linc02257-regulated gene expression was closely related to tumor development, such as collagen-containing extracellular matrix, organogenesis, activity of membrane protein receptors, and ion channel activity; the regulated signaling pathways were also closely related to tumors, such as neuroactive ligand-receptor interaction, the PI3K-AKT signaling pathway, calcium signaling pathway, and protein digestion and absorption, as shown in Figures 5,6.
Discussion
With the development of chip technology and high-throughput sequencing technology, more and more gene expression data are being generated. Most gene expression data are stored in public databases. Tumor RNA-seq data are mainly stored in TCGA database, which uses genome analysis technology of large-scale genome sequencing, with the purpose of mapping the complete genome of human cancer, improving understanding of the molecular basis of cancer pathogenesis, and improving the ability of cancer diagnosis, treatment, and prevention (14-16). The database of TCGA was established in 2006. The RNA-seq data of colon cancer tissues stored by TCGA contain rich gene expression content and have more comprehensive clinical information. Through the mining and analysis of TCGA data, the overview of lncRNA action during the occurrence and development of colon cancer can be found, its mechanism of action can be preliminarily understood, and lncRNA with clinical significance can be identified or screened concurrently.
As lncRNA is involved in various biological behaviors of cells, its abnormal expression is associated with a variety of diseases, which plays an important role in the occurrence and development of tumors. More and more studies have reported the role of LncRNA in tumor proliferation, cell invasion and migration, chemotherapy resistance, and stem cell ability in tumorigenesis and COAD progression (17,18). It has been reported that the abnormal expression of multiple lncRNAs is associated with the development of COAD, such as CYTOR, DDX11-AS1, UCA1, and CRNDE (19-22). However, there is still no comprehensive and in-depth study on the expression characteristics of lncRNA in COAD, and exploring the expression characteristics of lncRNA in CRC tissues has important significance for guiding experimental design.
In this study, we analyzed the RNA-seq data of COAD clinical samples in TCGA, including a total of 494 samples. The results showed that compared with normal control tissues, lncRNAs in COAD tissues had a large number of abnormal gene expression, with a total of 204 significant DEGs; 156 genes were up-regulated, and 48 genes were down-regulated. In this study, 10 genes with the most significant FDR were selected, and their |log2FC| was 3.61–9.00, which were Linc02418, Blacat1, ELFN1-AS1, CRNDE, AC002384.1, AL353801.1, LINC01645, AC073283.2, AC087379.1, and LINC00484. The abnormal expression of lncRNA is closely related to the occurrence and development of tumors. For example, in the study by Hongzhen et al. (23), the clinical data related to CRNDE were downloaded from both TCGA and the Gene Expression Omnibus (GEO). It was found that expression of CRNDE was abnormally increased in tumors, and the KM survival analysis plot revealed that it was closely related to the prognosis of cancer patients. In the study by Tian et al. (24), LINC02418 expression was found to be up-regulated in human colon cancer samples compared with adjacent tissues, and its high expression level was associated with poor prognosis of CRC patients and could be used as an indicator to predict prognosis. The results of another study (25) suggested that BLACAT1, as a cell cycle regulator, may be a potential target for the prevention and treatment of human colon cancer.
To investigate the effect of DEGs on the survival and prognosis of patients with colon cancer, we combined 204 lncRNA DEGs with 469 groups of survival data and obtained 5 DEGs that could affect the survival rate of patients using Cox analysis: Linc02257, Linc00891, Linc02474, Ac010789.1, and Ac083967.1. The survival curves of the other 4 genes were statistically significant except for the KM curve of Linc00891, that means the other 4 genes were significantly correlated with the prognosis of patients.
We selected the gene Linc02257 and performed functional GO enrichment analysis to predict its function. The results showed that Linc02257 is involved in a variety of biological behaviors, mainly related to tissue development, cell receptor expression, and substance metabolism, specifically including extracellular matrix of collagen, organogenesis, activity of membrane protein receptors, and ion channel activity, and these functional disorders are often related to tumor formation. Therefore, abnormal expression of Linc02257 is associated with tumor development. Further analysis of KEGG pathways of genes can reveal that Linc02257 is involved in tumor-related signaling pathway regulation, such as neuroactive ligand-receptor interaction, the PI3K-AKT signaling pathway, calcium signaling pathway, and protein digestion and absorption. In the study by Xiao et al. (26), it was also found that LINC02257 can function as an independent prognostic biomarker for COAD through the PI3K-AKT signaling pathway, and at the same time, LINC02257 may be a multifaceted and important immunotherapy-related enhancer RNA (eRNA) in different cancers.
Although we did not perform enrichment analysis for the other 3 genes that had a significant effect on the survival prognosis of patients, previous studies have shown that the other 3 genes are also closely related to the occurrence and development of colon cancer. The results of the study by Du et al. (27) showed that LINC02474 was associated with the prognosis of colon cancer patients: its down-regulation impaired the metastatic ability of CRC cells and promoted apoptosis, and up-regulation could promote the migration and invasion of CRC cells, while inhibiting apoptosis. The AC010789.1 gene may promote CRC progression by targeting the MicroRNA-432-3p/ZEB1 axis and the Wnt/β-catenin signaling pathway (28). The results of Wang et al. (29) showed that 15 genes related to colon cancer patient survival may play a key role in the regulation of cancer-related pathways as competitive endogenous RNAs (ceRNAs). At present, the regulatory mechanism of lncRNA is still not very clear, and the pathways regulated by different genes are also different; however, a large number of studies (30-32) have shown that the main mechanism of action is that lncRNA adsorbs miRNA as a ceRNA and then regulates the expression of target genes, that means the “lncRNA-miRNA-mRNA (coding RNA)” mechanism.
The data in TCGA database can basically reflect the changes of RNA gene expression. Analyzing the data can understand the expression characteristics of LncRNA and reveal its value in prognosis. However, TCGA data has some defects, because its data are provided by separate laboratories and the data are always changing, some recent tumor data have not been updated yet. And due to the influence of sequencing technology or sequencing quality, TCGA RNA-seq data cannot fully represent the expression of tumor genes. A large number of LncRNAs still are not been annotated, and therefore the recently discovered LncRNAs were not included in this analysis. In addition, the function and mechanism of several key LncRNAs screened in this study still need further experimental verification.
Conclusions
The data analysis in this study found 23 lncRNA genes with statistical significance for the prognosis of colon cancer tumors, and the survival rate analysis and enrichment analysis of 5 of them with statistical significance revealed that these genes were closely related to tumor tissue development, cell receptor expression, and substance metabolism, and the pathways regulated by different genes were different. There were still some shortcomings in this study: (I) because there are 23 genes related to the prognosis of colon cancer, we could not conduct KM curves of all 23 genes, and could not enrich and analyze 23 genes; we could only select a few; (II) TCGA data have some shortcomings: as their data are provided by various countries or laboratories, the data of some tumors in the recent past have not been updated, and due to sequencing technology or sequencing quality, RNA-seq data of TCGA do not fully represent the expression of tumor genes; (III) a large number of lncRNAs currently remain unannotated, and recently newly-discovered lncRNAs do not appear in this analysis. Therefore, the function and mechanism of action of several key lncRNAs screened in this study still need further experimental validation.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-384/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-384/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cappell MS. Pathophysiology, clinical presentation, and management of colon cancer. Gastroenterol Clin North Am 2008;37:1-24. v. [Crossref] [PubMed]
- Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer 2020;19:167. [Crossref] [PubMed]
- Zhou W, Zhang S, Li HB, et al. Development of Prognostic Indicator Based on Autophagy-Related lncRNA Analysis in Colon Adenocarcinoma. Biomed Res Int 2020;2020:9807918. [Crossref] [PubMed]
- Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer 2017;16:9. [Crossref] [PubMed]
- Zheng ZH, Wu DM, Fan SH, et al. Upregulation of miR-675-5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non-small cell lung cancer. J Cell Biochem 2019;120:18724-35. [Crossref] [PubMed]
- Tian F, Wang J, Zhang Z, et al. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol Res 2020;53:9. [Crossref] [PubMed]
- Zhao W, Geng D, Li S, et al. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med 2018;7:842-55. [Crossref] [PubMed]
- Zhi H, Lian J. LncRNA BDNF-AS suppresses colorectal cancer cell proliferation and migration by epigenetically repressing GSK-3β expression. Cell Biochem Funct 2019;37:340-7. [Crossref] [PubMed]
- Jelin AC, Vora N. Whole Exome Sequencing: Applications in Prenatal Genetics. Obstet Gynecol Clin North Am 2018;45:69-81. [Crossref] [PubMed]
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [Crossref] [PubMed]
- Ye S, Xu M, Zhu T, et al. Cytoglobin promotes sensitivity to ferroptosis by regulating p53-YAP1 axis in colon cancer cells. J Cell Mol Med 2021;25:3300-11. [Crossref] [PubMed]
- Lun AT, Chen Y, Smyth GK. It's DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol Biol 2016;1418:391-416. [Crossref] [PubMed]
- Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, et al. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer 2017;16:107. [Crossref] [PubMed]
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Travaglino A, Raffone A, Mascolo M, et al. TCGA Molecular Subgroups in Endometrial Undifferentiated/Dedifferentiated Carcinoma. Pathol Oncol Res 2020;26:1411-6. [Crossref] [PubMed]
- Xu M, Li Y, Li W, et al. Immune and Stroma Related Genes in Breast Cancer: A Comprehensive Analysis of Tumor Microenvironment Based on the Cancer Genome Atlas (TCGA) Database. Front Med (Lausanne) 2020;7:64. [Crossref] [PubMed]
- Poursheikhani A, Abbaszadegan MR, Nokhandani N, et al. Integration analysis of long non-coding RNA (lncRNA) role in tumorigenesis of colon adenocarcinoma. BMC Med Genomics 2020;13:108. [Crossref] [PubMed]
- Zhao B, Qu X, Lv X, et al. Construction and Characterization of a Synergistic lncRNA-miRNA Network Reveals a Crucial and Prognostic Role of lncRNAs in Colon Cancer. Front Genet 2020;11:572983. [Crossref] [PubMed]
- Yang J, Ma Q, Zhang M, et al. LncRNA CYTOR drives L-OHP resistance and facilitates the epithelial-mesenchymal transition of colon carcinoma cells via modulating miR-378a-5p/SERPINE1. Cell Cycle 2021;20:1415-30. [Crossref] [PubMed]
- Wan T, Zheng J, Yao R, et al. LncRNA DDX11-AS1 accelerates hepatocellular carcinoma progression via the miR-195-5p/MACC1 pathway. Ann Hepatol 2021;20:100258. [Crossref] [PubMed]
- Cui M, Chen M, Shen Z, et al. LncRNA-UCA1 modulates progression of colon cancer through regulating the miR-28-5p/HOXB3 axis. J Cell Biochem 2019; Epub ahead of print. [Crossref] [PubMed]
- Yang F, Li XF, Cheng LN, et al. Long non-coding RNA CRNDE promotes cell apoptosis by suppressing miR-495 in inflammatory bowel disease. Exp Cell Res 2019;382:111484. [Crossref] [PubMed]
- Hongzhen Z, Yanyu L, Xuexiang L, et al. The diagnostic and prognostic significance of long non-coding RNA CRNDE in pan-cancer based on TCGA, GEO and comprehensive meta-analysis. Pathol Res Pract 2019;215:256-64. [Crossref] [PubMed]
- Tian J, Cui P, Li Y, et al. LINC02418 promotes colon cancer progression by suppressing apoptosis via interaction with miR-34b-5p/BCL2 axis. Cancer Cell Int 2020;20:460. [Crossref] [PubMed]
- Su J, Zhang E, Han L, et al. Long noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15. Cell Death Dis 2017;8:e2665. [Crossref] [PubMed]
- Xiao J, Liu Y, Yi J, et al. LINC02257, an Enhancer RNA of Prognostic Value in Colon Adenocarcinoma, Correlates With Multi-Omics Immunotherapy-Related Analysis in 33 Cancers. Front Mol Biosci 2021;8:646786. [Crossref] [PubMed]
- Du T, Gao Q, Zhao Y, et al. Long Non-coding RNA LINC02474 Affects Metastasis and Apoptosis of Colorectal Cancer by Inhibiting the Expression of GZMB. Front Oncol 2021;11:651796. [Crossref] [PubMed]
- Duan W, Kong X, Li J, et al. LncRNA AC010789.1 Promotes Colorectal Cancer Progression by Targeting MicroRNA-432-3p/ZEB1 Axis and the Wnt/β-Catenin Signaling Pathway. Front Cell Dev Biol 2020;8:565355. [Crossref] [PubMed]
- Wang X, Zhou J, Xu M, et al. A 15-lncRNA signature predicts survival and functions as a ceRNA in patients with colorectal cancer. Cancer Manag Res 2018;10:5799-806. [Crossref] [PubMed]
- Guo Q, Cheng Y, Liang T, et al. Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression. Sci Rep 2015;5:17683. [Crossref] [PubMed]
- Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 2014;281:3766-75. [Crossref] [PubMed]
- Menges CW, Kadariya Y, Altomare D, et al. Tumor suppressor alterations cooperate to drive aggressive mesotheliomas with enriched cancer stem cells via a p53-miR-34a-c-Met axis. Cancer Res 2014;74:1261-71. [Crossref] [PubMed]



