Mutation profiles of synchronous colorectal cancers from a patient with Lynch syndrome suggest distinct oncogenic pathways
Introduction
Due to the high risk of synchronous and metachronous colorectal cancers (CRCs), the recommended treatment for CRC in patients with Lynch syndrome is colectomy (1). However, as the use of targeted therapies increases, including VEGF and EGFR inhibitors, an understanding of the molecular profiles of synchronous CRCs will be crucial in determining the most effective courses of treatment. If synchronous cancers are clonally derived, treatment may be relatively uniform. However, if they are distinct, each with a different oncogenic history, a treatment plan may need to include additional agents or therapeutic regimens.
In unselected patient populations with synchronous or metachronous CRCs, it is thought that different cancerous lesions in the same patient may follow distinct pathways of progression (2-4). Studies of matched synchronous CRCs suggested that exposure to different toxins at various locations within the colon were associated with distinct progression profiles (5,6). However, the effects of genetic background and environmental variation are confounding aspects in studying initiation and progression in sporadic cancers from different patients. Patients with Lynch syndrome frequently present with multiple synchronous or metachronous cancers (1,7,8). The presence of synchronous cancers arising in a single patient with a predisposing genetic mutation presents an opportunity to study the oncogenic pathways that lead to CRC while minimizing the effects of genetic background. In this study, we examined microsatellite instability (MSI) and genomic profiling in synchronous CRCs from a Lynch syndrome patient.
Materials and Methods
Pathology review and DNA extraction
Specimens were formaldehyde fixed, paraffin embedded and hematoxylin and eosin (H&E) stained. Representative sections were reviewed by a pathologist (CS) for histologic subtype, grade, staging and percent tumor burden. Sections were macrodissected to increase the representation of tumor DNA in the total DNA extracted from the specimen. DNA was isolated following cell lysis and proteinase K treatment using the QiaQuick extraction method (Qiagen, VHilden Germany).
Analysis of microsatellite instability (MSI)
DNA from normal and tumor specimens was amplified using the MSI analysis system version 1.2 (Promega, Madison WI, USA) according to the manufacturer’s instructions. Amplicons were detected using capillary electrophoresis on an ABI 3130xl Genetic Analyzer (Life Technologies, Carlsbad CA, USA) and the results were analyzed using GeneMapper V3.7 software (Life Technologies, Carlsbad CA, USA). The presence of instability in two, or more, of the five loci (>30%) was considered MSI-high (MSI-H).
SNaPshot mutation profiling
A SNaPshot single base extension assay was used to assess the mutation status of 62 loci in 7 genes (AKT1, BRAF, KRAS, NRAS, PIK3CA, PTEN and SMAD4) associated with CRC prognosis and treatment (9). SNaPshot products were separated using an ABI 3130xl Genetic Analyzer (Life Technologies, Carlsbad CA, USA) and compared to positive and normal controls for interpretation.
Massively parallel DNA sequencing
Multiplex amplicon-based sequencing libraries were prepared using the GeneRead DNA-seq Human Comprehensive Cancer Panel NGHS-501X (Qiagen, Hilden Germany) following the manufacturer’s instructions. This panel targets coding and UTR regions of 124 commonly mutated genes in multiple cancer types. Once prepared, libraries were sequenced using a MiSeq 300 cycle V2 reagent kit (Illumina, San Diego CA, USA) with MiSeq Control software V2.3.0.3 and RTA software V1.18.42.0. Variant analysis was performed using the CLCbio genomics workbench (Qiagen, Hilden Germany).
Results
An 81-year-old Egyptian male presented with weight loss, upper quadrant abdominal and rectal pain and blood streaked stools. The medical history was significant for cancer including a tumor of unreported origin removed by a partial small bowel resection in his 50’s, renal cell carcinoma removed by nephrectomy in his 60’s and prostate cancer treated with implantation of radioactive seeds in his 70’s. The family history was also significant for cancer. Two first-degree relatives had colon cancer, 2 first-degree relatives had kidney cancer and a first-degree relative had an unspecified lymph node/head and neck cancer.
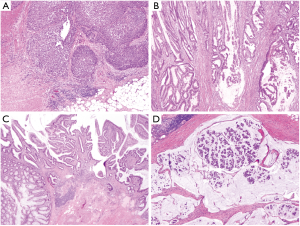
After total proctocolectomy, 6 lesions were identified. These included 5 colonic and 1 rectal lesion. Lesion 1 (medullary carcinoma, 5 cm in greatest dimension, T4N0M0) was located in the left colon near to the splenic flexure (Figure 1A). Lesion 2 (moderately differentiated invasive adenocarcinoma, 6 cm, T3N0M0) was located in the right colon near to the hepatic flexure (Figure 1B). The colon also contained 3 early invasive carcinomas (T1N0M0) arising from tubulovillous adenomas that, from proximal to distal, were 1cm (lesion 3, Figure 1C), 1.3 cm (lesion 4) and 1.2 cm (lesion 5). Lesion 6 was identified in the rectum (invasive mucinous adenocarcinoma with clusters of signet ring cells, 6.5 cm, 8 of 15 lymph nodes involved, T3N2bM0) (Figure 1D).
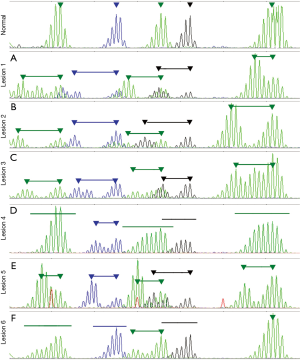
MSI was detected in all lesions. However, the character of MSI was different in each (Figure 2). Differences included the number of unstable loci and the pattern and extent of instability. Each lesion was also screened for a panel of 62 hot-spot variants in 7 genes related to prognosis and treatment in colon cancer (9). BRAF V600E was not detected in any of the lesions (data not shown) suggesting Lynch syndrome rather than sporadic CRC (10). Other identified variants included KRAS G12D in lesion 1 and KRAS G13D in lesion 6 (data not shown). To identify potential Lynch syndrome-related germline variants, MLH1, MSH2, MSH6 and PMS2 were examined in DNA isolated from peripheral blood. A frameshift variant was detected in MSH2, c.2082delT (p.Phe694Leufs*16). This variant was previously reported in patients with CRC and established a diagnosis of Lynch syndrome in this patient (11). Taken together, these findings suggest that each lesion arose from a unique event in a background predisposing to cancer.
To understand better how these cancers developed, sequencing of 124 cancer-related genes was performed. Only lesions 1, 2 and 3 had an adequate amount of high quality DNA for this analysis. The majority of variants (48.7%, 186/382) including substitutions, insertions and deletions were identified in all 3 lesions. In the absence of normal DNA to use for comparison, the common variants are a good estimate of the patient’s germline variation. However, some of these variants may also be hot-spot mutations that occurred independently in each lesion. Germline variants have an expected frequency of approximately 50% (heterozygous) or 100% (homozygous). Of the presumed germline variants identified in the patient’s CRCs, 89.2% have a variant frequency that is indicative of a heterozygous (58.6% with a variant frequency of 40–60%) or homozygous (30.6% with a variant frequency of 90–100%) state. Presumed germline variants were found in 61 genes, all with low or moderate impact predictions (Tables S1,S2). The single high impact variant observed in all lesions was the previously identified c.2082delT in the MSH2 gene.
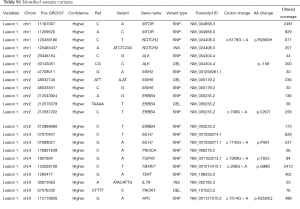
Full table

Full table
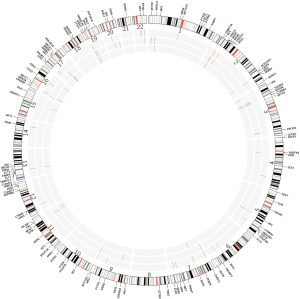
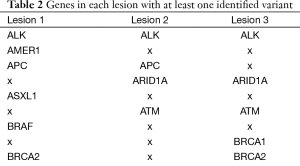
To determine the lesion-specific variants, the presumed germline variants were subtracted from each lesion. After subtraction, the percent of variants in each lesion that met the criteria for homozygous or heterozygous state was drastically reduced: 8.1% in lesion 1, 11.8% in lesion 2 and 2.2% in lesion 3. These values are significantly different from the values in the presumed germline variation suggesting that the majority of variants ascribed to the lesions are the result of somatic mutation. Respectively, lesions 1, 2 and 3 had 96, 74 and 46 somatic variants with similar distributions of types (Table 1). To compare the mutational landscape between lesions, similarities and differences in affected genes were examined (Figure 3, Table 2). All 3 lesions had somatic variants in two genes, SMARCA4 and ALK. For SMARCA4, each lesion contained distinct high impact variants: lesion 1 had a non-synonymous coding variant, lesion 2 had a frameshift and lesion 3 had two distinct non-synonymous coding variants. In ALK, lesion 2 contained a C-terminal deletion while lesions 1 and 3 contained the same N-terminal deletion. These data suggest an important role for loss of function of these genes in the development of these lesions. Additionally, as expected, all 3 lesions had different combinations of variants in WNT pathway genes including APC, ARID1A, CTNNB1, and FBXW7.
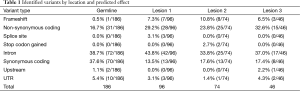
Full table
Full table
There were also significant differences between the lesions. Thirteen genes had variants in 2 lesions and 34 genes had variants in only a single lesion indicating significant differences between lesions. Lesion 1 had variants in 18 unique genes involved in Notch, RAS, and NF2 signaling (Table 2). In addition, a variant in MSH2 (c.2634+1G>A) is described as pathogenic in ClinVar because it interrupts a canonical splice site and is likely the second MSH2 inactivating event (13). Lesion 2 had variants in 11 unique genes involved in the EGFR, KIT, MTOR and SRC signaling pathways and transcriptional regulation (CREBBP, DNMT3A, MYD88, PAX5) (Table 2). Of note, a frameshift mutation was identified in MSH6 (c.3205delG, p.G1070fs*9). Heterozygosity of both partners in the MSH2/MSH6 heterodimer may result in reduced function. Lesion 3 was the least advanced of the cancers studied (T1N0M0) and had the smallest number genes containing variants. Identified variants did not correspond to obvious signaling molecules, but were present in genes involved in DNA repair, gene expression and proteasome function. These data, like the MSI data, indicate distinct oncogenic histories for each lesion.
Discussion
The presence of multiple synchronous cancers in this patient allowed for a unique analysis of genetic diversity among CRCs without the confounding effects of genetic background. Taken together, these results provide a diverse picture of colon carcinogenesis with distinct histology, MSI patterns and gene mutation profiles.
Two genes had variants identified in all 3 sequenced lesions. Both SMARCA4 and ALK are involved in other cancer types, but neither was identified at significant levels in the survey of CRCs performed by The Cancer Genome Atlas (14). SMARCA4 encodes a subunit of the SWI/SNF complex and mutations could alter transcriptional regulation through that mechanism (15). Outside of childhood neuroblastoma, ALK mutations have not been widely identified, although ALK fusions are involved in the development of multiple cancer types (16). It is possible that the development of variants in these genes is unique to this patient or to patients with Lynch syndrome. Comprehensive DNA sequencing studies of cancers in patients with Lynch syndrome are needed to identify genes that may be particularly susceptible to mutation in this patient population.
The analysis of MSI is usually focused on the interpretation: stable, low or high. However, when studying the relatedness of cancerous lesions, the pattern of errors observed can act as a genetic fingerprint. This is because MSI assesses the accumulation of replication errors during the clonal expansion of cancer cells. Therefore, independent cancers should have different patterns of errors owing to their unique oncogenic history and the randomness of the errors. In this study, each synchronous lesion had a distinct pattern of errors indicating distinct oncogenic histories. The genetic differences implied by the MSI analyses were observed in the SNaPshot and sequencing data as each lesion had a unique set of variants affecting a unique set of genes. Therefore, it may be possible to use MSI analysis as a screening tool to determine the relatedness of synchronous of metastatic lesions.
It is estimated that approximately 4% of unselected patients presenting with CRCs have a synchronous cancer and the 5-year survival rate for both synchronous and single lesion CRC was approximately 50% (17,18). These studies, while comprehensive, examined data from patients collected up until 2004 prior to the wide-spread use of targeted agents in CRC therapy. Targeted therapies have shown positive effects on progression free and overall survival, but the presence of multiple cancers complicates the choice of targeted therapies (19). In the current study, 2 of 6 lesions harbored activating KRAS mutations and all 3 sequenced lesions had either EGFR, ERBB2 or ERBB4 variants identified. EGFR antibody therapy is unlikely to be effective for the lesions with KRAS mutations and the variants in ERBB2 and ERBB4 may affect the efficiency of agents targeted to those molecules. These findings suggest that for patients with synchronous cancers, surgical resection may still be the best option as each tumor can have a different response to targeted agents. Further, when targeted therapies are considered, multiple lesions should be profiled, to reveal the complexity of disease and to optimize treatment.
Acknowledgements
Funding: This work was supported by The National Institutes of Health and National Cancer Institute (P50CA095103) to C Shi. The authors would like to acknowledge technical assistance from Travis Clark, PhD; Thomas Stricker, MD, PhD and Vanderbilt’s VANTAGE genomics core facility, which is supported by the Clinical and Translation Science Award (5UL1 RR024975-03), the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126) and the National Institutes of Health and The National Center for Research Resources (G20 RR030956).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Schlussel AT, Gagliano RA Jr, Seto-Donlon S, et al. The evolution of colorectal cancer genetics-Part 2: clinical implications and applications. J Gastrointest Oncol 2014;5:336-44.
- Aslanian HR, Burgart LJ, Harrington JJ, et al. Altered DNA mismatch repair expression in synchronous and metachronous colorectal cancers. Clin Gastroenterol Hepatol 2008;6:1385-8.
- Lawes DA, Pearson T, Sengupta S, et al. The role of MLH1, MSH2 and MSH6 in the development of multiple colorectal cancers. Br J Cancer 2005;93:472-7.
- Bae JM, Cho NY, Kim TY, et al. Clinicopathologic and molecular characteristics of synchronous colorectal cancers: heterogeneity of clinical outcome depending on microsatellite instability status of individual tumors. Dis Colon Rectum 2012;55:181-90.
- Zauber P, Huang J, Sabbath-Solitare M, et al. Similarities of molecular genetic changes in synchronous and metachronous colorectal cancers are limited and related to the cancers' proximities to each other. J Mol Diagn 2013;15:652-60.
- Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012;61:847-54.
- Fitzgibbons RJ Jr, Lynch HT, Stanislav GV, et al. Recognition and treatment of patients with hereditary nonpolyposis colon cancer (Lynch syndromes I and II). Ann Surg 1987;206:289-95.
- Box JC, Rodriguez-Bigas MA, Weber TK, et al. Clinical implications of multiple colorectal carcinomas in hereditary nonpolyposis colorectal carcinoma. Dis Colon Rectum 1999;42:717-21.
- Mikhitarian K, Pollen M, Zhao Z, et al. Epidermal growth factor receptor signaling pathway is frequently altered in ampullary carcinoma at protein and genetic levels. Mod Pathol 2014;27:665-74.
- Cushman-Vokoun AM, Stover DG, Zhao Z, et al. Clinical utility of KRAS and BRAF mutations in a cohort of patients with colorectal neoplasms submitted for microsatellite instability testing. Clin Colorectal Cancer 2013;12:168-78.
- Wu Y, Nyström-Lahti M, Osinga J, et al. MSH2 and MLH1 mutations in sporadic replication error-positive colorectal carcinoma as assessed by two-dimensional DNA electrophoresis. Genes Chromosomes Cancer 1997;18:269-78.
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92.
- Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980-5.
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7.
- Roskoski R Jr. Anaplastic lymphoma kinase (ALK): structure, oncogenic activation, and pharmacological inhibition. Pharmacol Res 2013;68:68-94.
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011;11:481-92.
- Latournerie M, Jooste V, Cottet V, et al. Epidemiology and prognosis of synchronous colorectal cancers. Br J Surg 2008;95:1528-33.
- Lam AK, Carmichael R, Gertraud Buettner P, et al. Clinicopathological significance of synchronous carcinoma in colorectal cancer. Am J Surg 2011;202:39-44.
- Luu C, Arrington AK, Schoellhammer HF, et al. Targeted therapies in colorectal cancer: surgical considerations. J Gastrointest Oncol 2013;4:328-36.


