
Comprehension of rectosigmoid junction cancer molecular features by comparison to the rectum or sigmoid colon cancer
Highlight box
Key findings
• We showed the distinctive molecular profiles of the sigmoid colon, rectosigmoid junction, and rectum. We observed a gradual change in the key genes of CRC along the bowel and higher TGF-β pathway alterations in the rectosigmoid junction, and rectum.
What is known and what is new?
• Carcinomas of the rectosigmoid junction are frequent; however, specific data on these tumors are sparse, as they are frequently assigned to either the colon or rectum.
• The next-generation sequencing (NGS) data of the patients were analyzed to study the molecular characteristics of the carcinomas in different locations of the bowel.
What is the implication, and what should change now?
• Our results may contribute to the selection of individualized treatment for tumors at different locations.
Introduction
Up to 10% of colorectal carcinomas are adenocarcinomas of the rectosigmoid junction (1,2). The rectosigmoid junction (ICD-O; C-19) is encoded as a separate segment of the large intestine under the Classification of Disorders for Oncology, International Classification of Diseases for Oncology (ICD-O), 3rd Edition of the World Health Organization (www.who.int). There used to be no international consensus definition for the rectum. The most commonly definitions of the proximal extent of the rectum were 15 cm from the anal verge and the sacral promontory. The ‘‘sigmoid take-off’’ as a more consistent and accurate classification of rectal versus sigmoid cancers—an anatomic, image-based definition of the junction of the mesorectum and mesocolon—has emerged as the consensus of international experts (3).
In most studies on colorectal carcinomas, the rectosigmoid junction has not been evaluated separately but has been considered part of the rectum (4,5) or colon (6). To the best of our knowledge, only a few studies have sought to analyze adenocarcinomas of the rectosigmoid junction and to compared the region to the adjacent colorectal segments to examine the tumor characteristics of each (7). GLOBOCAN showed that colorectal ranked third in terms of incidence, but second in terms of mortality worldwide in 2020 (8). Tumors were classified as left-sided colon cancer (LCC), if they were found in the splenic flexure up to the rectum, including descending, and sigmoid and/or rectosigmoid cancers, and were classified as right-sided colon cancer (RCC) if they were found in the caecum, ascending or transverse colon. LCCs have higher incidence rates than RCCs in global (9). A review of pathological and autopsy records of 5,817 patients diagnosed found that liver metastases are more commonly found in LCCs due to its anatomical situation with regard to portal circulation (10). Due to the fact that RCCs are more frequently diploid and characterized by v-raf murine sarcoma viral oncogene homolog B1 (BRAF) mutations, mucinous histology, high microsatellite instability, and CpG island methylation, whereas LCCs were found to have frequently p53 and Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations, it was discovered in in a systematic review and meta-analysis included more than 1.4 million patients that an absolute 19% lower risk of death was found to be significantly associated with having a tumor that originated on the left side of the colon (11). For example, BRAF- V600E mutant CRLM (mutation in a specific BRAF locus V600E) is associated with a poor prognosis (12). The pattern of lymphatic spread of the rectosigmoid junction differs to that of the sigmoid or rectum (13). The approach to treating colorectal cancers (CRCs), however, has evolved to be more differentiated and individualized (14). For the early diagnosis, prognosis, and treatment of rectosigmoid junctional carcinoma, it is therefore critical to identify effective potential molecular biomarkers. With the development of next-generation sequencing (NGS) technology, we can comprehensively understand the molecular features of CRC. Retrospective analyses of multiple trials have shown that CRC patients with RAS/BRAF wild-type benefit from anti-epidermal growth factor receptor (EGFR) therapy (15,16). CRCs are a heterogeneous group of diseases with complex genetic and epigenetic alterations (17). The molecular classification of such diseases is thus increasingly important in clinical decision making (18).
In a recent study, it was shown that the frequencies of the CpG island methylator phenotype (CIMP), microsatellite instability-high (MSI-H), and BRAF mutations in cancer progressively increase from the rectum to the ascending colon along the colorectum subsites (19). A previous study also revealed that the sigmoid-rectal region appears to have unique molecular features compared to those of other colon-sided locations (20). The distinctive competing endogenous RNA and long non-coding RNAs of the rectosigmoid junction cancer have been reported (21,22). A multi-omics study of gastric cancer has also demonstrated the heterogeneity of molecular features (23). Thus, differences in disease prognosis and progression urgently need to be understood to help in the identification of exclusive biomarkers for the colon, rectum, and rectosigmoid junction.
In this study, we retrospectively reviewed all the clinical and NGS panel data of 96 CRC patients. We also summarized the molecular alterations based on the tumor location of the sigmoid colon, rectosigmoid junction, and rectum. Finally, we compared the molecular features of colon, rectum, and rectosigmoid junction cancer. We present this article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-120/rc).
Methods
Study design and patients
The data of 96 CRC patients treated at the Wuxi Hospital Affiliated to the Nanjing University of Chinese Medicine from January 2017 to December 2021 were retrospectively analyzed. Patients older than 18 years of age were diagnosed with carcinomas in the sigmoid colon, rectosigmoid junction or rectum; and genetic testing information of tissue or tissue samples available for genetic testing were included. Patients younger than 18 years of age, without genetic testing information or tissue available, or those who declined informed consent were excluded. Classification of tumors would be based on their anatomical location:Sigmoid: distal sigmoid tumors that arise above the sigmoid take-off; Rectosigmoid: tumors that straddle the take-off; Rectal: high/upper third rectal tumors which are located below the sigmoid take-off, but above the peritoneal reflection.
All the patients underwent NGS by the 1021-gene panel at Geneplus-Beijing (Beijing, China). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Wuxi Hospital Affiliated to Nanjing University of Chinese Medicine (No. 201809001J01-01), and each patient provided informed consent.
DNA sequencing
Fresh tissues or formalin-fixed paraffin-embedded tissues and 10 mL of matched peripheral blood were obtained from each patient for matched tumor-normal NGS testing. As previously described (24), the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) was used for the tissue sample extraction. Comprehensive genomic profiling was performed using a custom-designed NGS panel containing 1,021 cancer-associated genes (Table S1) and the genomic DNA sequencing libraries were prepared in accordance with the instructions of the KAPA DNA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA). DNA sequencing was performed on a DNBSEQ-T7RS sequencer (MGI Tech, Shenzhen, China) or Gene + Seq-2000 sequencing system (GenePlus-Suzhou, Suzhou, China) with a 100-bp paired-end configuration. The reads were aligned to the human genome build GRCh37 using a Burrows-Wheeler aligner (25). MuTect2 (3.4-46-gbc02625) (26) was used to call single nucleotide variants (SNVs), while GATK (The Genome Analysis Toolkit) was employed to call small insertions and deletions (indels). All the final candidate variants were verified with the integrative genomics viewer browser. The tumor mutation burden (TMB) was calculated as the number of somatic non-synonymous SNVs and indels per Mb in the coding region, with a variant allele fraction of ≥0.03. The MSI status was inferred using MSIsensor (v.0.5) software (37-ming), and MSI-H (MSI-high) was defined as a cut-off MSI score >8%.
To premise of successful and accurate sequencing, a process of quality control was compulsory. The following factors were employed in this study to assess the quality of genetic sequencing: library complexity, insert size, median depth, Q20 ratio, and Q30 ratio. The library complexity represents the sample size of all input samples eventually incorporated in the library and sequenced. DNA degradation is measured using insert size, with a lower value indicating more DNA degradation. The Q30 ratio, which measures the percentage of reads with a sequencing accuracy of more than 99.9%, and the Q20 ratio, which measures the percentage of reads with a sequencing accuracy of more than 99%, are two metrics that reflect the quality of genetic sequencing. The criteria utilized in this work were 20%, 150 bp, 500 X, 90%, and 80%, respectively, for library complexity, insert size, median depth, Q20 ratio, and Q30 ratio.
Statistical analyses
The difference in patient demographics was evaluated using the Fisher t-test. Data on smoking which missed were not included in the statistical analyses. GraphPad Prism 8.0.2 (GraphPad Software, Inc.) was used to perform the other statistical analyses. A 2-tailed unpaired Mann-Whitney U test was used to evaluate the differences. Results with P values of less than 0.05 were regarded as statistically significant.
Results
The clinicopathologic characteristics of the patients in our cohort
A total of 96 patients met the study requirements and were enrolled in this study. Information about the location of the primary tumor was available for the entire cohort. Table 1 summarizes the baseline characteristics of the participants stratified by tumor location. Median age, mismatch repair (MMR) status, disease stage, metastatic location, histology, and family history all differed based on the tumor location, while the differences were not statistically significant. The patients with rectum cancer were slightly older than those with rectosigmoid junction cancer or sigmoid colon cancer. In each group, >60% patients were in the advanced stage. Almost all of the patients had adenocarcinoma and proficient MMR (pMMR). A higher proportion of lung metastasis was found in the patients with rectum cancer than those with rectosigmoid junction cancer or sigmoid colon cancer (25.7% vs. 12.1% vs. 14.3%, P=0.1565); however, in relation to distal metastasis, there were no significant differences between the three groups. Information on the family history of cancer was available for 58 patients, and 17 of the 58 patients had at least 1 family member who had a history of cancer, including 6 who had a family history of CRC and 5 who had a family history of gastrointestinal cancer.
Table 1
| Clinical feature | Sigmoid colon (n=33) | Rectosigmoid junction (n=28) | Rectum (n=35) | P |
|---|---|---|---|---|
| Median age (years) | 55 [38–76] | 52 [29–70] | 60 [29–76] | 0.23 |
| Gender | 0.46 | |||
| Female | 13 (39.4) | 15 (53.6) | 14 (40.0) | |
| Male | 20 (60.6) | 13 (46.4) | 21 (60.0) | |
| Disease stage | 0.23 | |||
| II/III | 10 (30.3) | 6 (21.4) | 14 (40.0) | |
| IV | 23 (69.7) | 22 (78.6) | 21 (60.0) | |
| Metastatic location | 0.23 | |||
| Liver | 12 (36.4) | 10 (35.7) | 8 (22.9) | |
| Lung | 4 (12.1) | 4 (14.3) | 9 (25.7) | |
| Peritoneum | 4 (12.1) | 1 (3.6) | 1 (2.9) | |
| Others | 1 (3.0) | 3 (10.7) | 1 (2.9) | |
| MMR status | 0.53 | |||
| dMMR | 0 | 1 (3.6) | 1 (2.9) | |
| pMMR | 33 (100.0) | 27 (96.4) | 34 (97.1) | |
| Histology | 0.53 | |||
| Adenocarcinoma | 33 (100.0) | 27 (96.4) | 34 (97.1) | |
| Signet/mucinous | 0 | 1 (3.6) | 1 (2.9) | |
| Family history of cancer | 0.42 | |||
| Absent | 16 (48.5) | 17 (60.7) | 8 (22.9) | |
| Present | 6 (18.2) | 5 (17.9) | 6 (17.1) |
Data are presented as No. (%) or median [range]. Differences in categorical baseline characteristics were compared using the χ2 test. CRC, colorectal cancer; dMMR, mismatch repair-deficient; pMMR, mismatch repair-proficient.
Molecular characteristics of the patients with different tumor locations
We analyzed the molecular characteristics of the 96 samples in terms of the different tumor locations. The Tumor Protein P53 (TP53), adenomatous polyposis coli (APC), KRAS genes were frequently mutated in our cohort, and had mutation frequency rates of 82.3%, 76.1%, and 43.8%, respectively. The next most frequently mutated genes were phosphoenolpyruvate carboxykinase 1 (PCK1) (27%) and B-cell lymphoma-2 like 1 (BCL2L1) (27%) in the sigmoid colon, v-myc avian myelocytomatosis viral oncogene homolog (MYC) (29%) and SMAD Family Member 4 (SMAD4) (25%) in the rectosigmoid junction, and F-box and WD repeat domain containing 7 (FBXW7) (31%) and Transcription factor 7-like 2 (TCF7L2) (29%) in the rectum (Figure 1A).

An analysis was also conducted to examine the co-occurrence of the mutated genes (Figure S1). In the rectum group, the TP53 and KRAS mutation were mutually exclusive (P<0.05), and the FBXW7 and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutations showed co-occurrence (P<0.05). In the rectosigmoid junction group, the KRAS and TP53 mutations were mutually exclusive, but no statistically significant difference, as were the TP53 and PIK3CA mutations (P<0.05). Figure 1B shows the proportions of the key genes in the different tumor location groups. The rates of the KRAS, neuroblastoma RAS viral oncogene homolog (NRAS) and PIK3CA mutations increased, moving distally, while the rates of APC and BRAF decreased. The prevalence of TP53 mutations was similar at different tumor locations. KRAS and NRAS have been grouped together. The prevalence of the RAS mutation was significantly higher in the rectum group than the other two groups (P=0.03).
The RAS gene subtypes were further analyzed (Table S2). KRAS p.G12D, p.G12V, and p.G13D were the common subtypes. KRAS p.G12C was only detected in a few cases of sigmoid and rectosigmoid junction carcinomas. NRAS mutations were mainly detected in rectum cancer, and NRAS p.Q61K and p.G12D were the common subtypes. The two patients with deficient-MMR were also examined for MSI-H. Both patients carried the germline mutations of PMS1 Homolog 2, Mismatch Repair System Component (PMS2) and MutS Homolog 6 (MSH6), and 1 patient had a family history of gastrointestinal tumors. The TMB was also analyzed. The median TMBs were 6.96, 4.80, and 6.00 mut/Mb for sigmoid colon, rectosigmoid junction, and rectum cancer groups, respectively. Overall, we observed a gradual distribution of key genes along the sigmoid colon to the rectum.
Distinctive molecular profiles and pathway enriched of different tumor locations
To explore the distinctive molecular profiles of the different tumor locations, we investigated the difference in genomic variations between the different groups. As Figure 2 shows, there were almost no significant molecular differences among the three groups. The results of the comparison showed that the prevalence of the fms-related tyrosine kinase 3 (FLT3), fms-related tyrosine kinase 1 (FLT1), and PCK1 mutations was significantly higher in the sigmoid colon than the rectosigmoid junction (P=0.0057, P=0.031, P=0.049). The prevalence of the FLT3, FLT1, and PCK1 mutations was also higher in the rectum than the rectosigmoid junction, but the difference was not significant. As mentioned above, there were some differences (P>0.05) in the prevalence of the key mutated genes between the sigmoid colon and rectum. the FLT3, FLT1, and PCK1 genes were in the top 10 in our cohort and were particularly prevalent in the sigmoid colon and rectum cancer patients. We also analyzed the mutations of FLT1, FLT3, and PCK1 in 5,050 CRC patients that assessed as a CRC group regardless of site through cBioportal (https://www.cbioportal.org/), and the incidence rates of the FLT1, FLT3, and PCK1 mutations were 7%, 6%, and 5%, respectively.

The systemic characterization of the genomic alterations into signaling pathways will help us to further understand the molecular characteristics of different tumor locations. All the genes defined as cancer genome maps pan-cancer analysis project mutations have been assigned to 10 signaling pathways (27). As Figure 3A shows, the common pathways were MYC, TP53, transforming growth factor beta (TGF-β), Wingless-Type MMTV Integration Site Family (WNT), Notch, phosphoinositide 3-kinase (PI3K), and receptor tyrosine kinases (RTK)-RAS. Almost no differences were observed between the three groups in terms of the 10 pathways, but there was a higher proportion of cell-cycle alterations in the sigmoid colon than the rectosigmoid junction (30.3% vs. 3.6%, P<0.001). Meanwhile, a higher proportion of MYC pathway was observed in the rectosigmoid junction than that in rectum and sigmoid colon (28.6% vs. 15.2% vs. 17.1%, P=0.278, P=0.202, P=0.171); a higher proportion of TGF-β pathway was also observed in the rectosigmoid junction, and rectum than the sigmoid colon (39.3% vs. 34.3% vs. 18.2%, P=0.121, P=0.067, P=0.682). Previous studies have shown that TGF-β signaling, which does not respond to immune checkpoint inhibitors (ICIs), is significantly increased in urothelial cancer, breast cancer, and others (28,29). Thus, to investigate the correlations between the different tumor locations, gene mutations were also assigned to immune-related signaling pathways as defined by a previous study (30). With the exception of a higher proportion of activated dendritic cells being found in sigmoid colon cancer than in rectosigmoid junction cancer, or rectum cancer (27.3% vs. 10.7% vs. 5.7%, respectively, P=0.03), no difference was observed among the three groups across the immune-related signaling pathways (Figure 3B). Overall, tumors at different locations showed distinct molecular profiles and pathways, while the absence of significant differences also supports molecular gradients across locations.

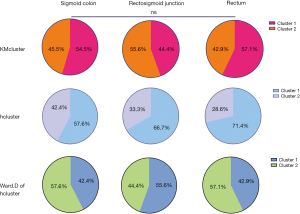
Clustering by molecular profiles and comparisons of different tumor locations
We also explored whether there were differences in clusters based on the molecular profiles between the tumor locations. In the analysis, 3 methods were employed for the clustering; that is, the K-mean cluster (KMcluster), maximum of hierarchical cluster (hcluster), and Ward.D of hcluster. The optimal number of clusters derived by each of these methods was 2 (Figure S2). When the sigmoid colon, rectosigmoid junction, and rectum were compared to each other, they did not differ based on the results of either method of clustering (Figure 4).
We also explored the mutational signatures of our cohort. The common mutational signatures in our cohort were 8, 12, 17, 20, 22, and 28. Signatures 20 and 22 are believed to be associated with DNA MMR and with defective DNA MMR, respectively. Notably, the proportion of Signature 22 was higher in the group of rectum cancer than the other two groups (82.9% vs. 66.7% vs. 57.1%, respectively, P=0.08). We also clustered the mutational signatures of our cohort by KMcluster. The optimal number of clusters given was 2, and no difference was found among the different tumor locations (Figure S3).
Discussion
CRC is one of the most prevalent and lethal malignancies in the world. Given that at this moment the molecular characteristics of the tumor and the tumor’s location directly impact medical therapy. Rectosigmoid junction tumors are either treated as rectum tumors or sigmoid colon tumors due to their heterogeneous characteristics. Thus, exploring the molecular characteristics of the rectosigmoid junction could extend understandings of this type of tumor and guide the selection of treatments. In this study, we analyzed the key genes and compared the molecular characteristics between the different tumor locations from the sigmoid colon to the rectum. We also assessed the relevant pathways and clusters to examine any differences related to the different tumor locations.
As is well known, NGS platforms have made it possible to massively parallelize the high-throughput sequencing of millions to billions of DNA fragments. Contrast this with single DNA sequences performed using first-generation Sanger sequencing, which would miss certain variants, such as tiny insertion/deletion mutations. A major advantage of NGS compared with real-time polymerase chain reaction (PCR) is that target-specific primer is not required. NGS can also generate sequences of numerous molecular in one sequencing run, and enabled the inquiry of nearly every base in the genes. Besides, the decrease in instrumentation and the running costs of NGS makes it more suitable for clinical usage. Therefore, the NGS-based inquiries required for less hypothesis driven and examine all genes, the cost is less expensive and the data were more rapidly obtained, is helpful for further exploration on the various molecular features of rectosigmoid junction cancer and applicable to the further gene-therapy for patients (31).
We examined the molecular characterization of the 96 patients with CRC based on the tumor locations and the molecular feature changes along the bowel in the distal colon. There were no differences in the clinicopathologic characteristics of the three groups. Similar to previous studies that have reported that rectum patients have a high ratio of lung metastasis (32,33), the patients in the rectum group in this study also had a higher ratio of lung metastasis than those in the other groups; however, the difference was not significant. Notably, we found that the molecular characteristics between the different tumor locations were similar but distinct. The top 3 mutated genes were consistent in all groups. However, for most of the key genes, we found that the proportions changed gradually with the tumor locations. Specific alterations of note included a decrease in the BRAF V600 mutation from the sigmoid colon to the rectum, and an increase in the PIK3CA and RAS mutations from the sigmoid colon to the rectum. We also found a significantly higher incidence of RAS mutations in the rectum cancer group than the others. Recent studies have assessed changes in the molecular features along the bowel and reported the same molecular trend (19,20), but no differences in RAS mutations were previously reported in the sigmoid colon to the rectum.
In relation to the molecular profiles across the 3 locations, no difference was found. However, the different tumor locations had distinctive molecular profiles. FLT1, FLT3, and PCK1 were less common in the rectosigmoid junction group than the other groups. FLT1 and FLT3 are members of the vascular endothelial growth factor receptor family, and these genes are the target genes of Bevacizumab (34). The 3rd edition of the ICD-O states that the rectosigmoid junction should now be classified as 1 independent segment of the large intestine (ICD-O; C-19), rather than as part of the colon (ICD-O; C-18) or rectum (ICD-O; C-20). Therapy for cancers at the rectosigmoid junction should differ to that for cancers at the sigmoid colon and rectum given its special location (7). However, the treatment of rectosigmoid junction is more comply with the treatment of rectal or colon cancer (35).
Our study showed that the rectosigmoid junction has a distinctive molecular profile, and the rectosigmoid junction should be considered independently and cannot be assigned to the sigmoid colon or the upper rectum. Recent studies have also revealed differences between the three locations. Park et al. reported that the clinicopathological characteristics of the rectosigmoid junction cancer are similar to those of sigmoid or rectal cancer, but the rectosigmoid junction cancer has different patterns of lymphatic spread compared to the sigmoid colon or rectum cancer and more frequently metastasizes to the pararectal nodes (13). The distinctive RNA network of the rectosigmoid junction has also been reported (21,22). A recent study reported that the rectosigmoid junction had a deviant behavioral pattern compared to the patterns of adjacent bowel segments, including lower 5-year overall survival and higher lymph vascular invasion (35). Thus, individualized treatment strategies urgently need to be established for the rectosigmoid junction.
The distinctive molecular profiles of the three locations were also examined in terms of the signaling pathways. The TGF-β signaling pathway was more highly expressed in the rectosigmoid junction, and rectum. The TGF-β signaling pathway regulates tissue development and homeostasis, and genomic alterations in this signaling pathway are involved in CRC progression (36,37). Genomic TGF-β pathway alterations have been identified in 30% of rectosigmoid junction or rectum cancer patients and only 12% of in sigmoid colon cancer patients (38). Research using The Cancer Genome Atlas (TCGA) and Memorial Sloan Kettering Cancer Center data sets have shown that alterations in the TGF-β pathway are correlated with worse overall survival in patients with metastatic CRC (39,40). Notably, shorter overall survival is associated with an altered TGF-β pathway, which is positively associated with those who receive a first-line treatment with an anti-EGFR antibody; however, sadly there is no evidence of an association between altered of TGF-β pathway and treatment in patients receiving an anti-vascular endothelial growth factor antibody. Some pre-clinical studies have shown that the TGF-β pathway achieves anti-EGFR therapy resistance by either protein kinase B (AKT) activation or SMAD4-associated epithelial-mesenchymal transformation (41,42). In kras-mutated/trp53-deleted murine colonocytes, either Myc activation or TGF-β inactivation increased tumor sizes, furthermore in human CRC, gain-of-function alterations in Myc and loss-of-function alterations in TGF-β exhibit, for example in SMAD2/3/4 genes, a masking epistatic interaction and are functionally redundant (43). Moreover, TGF-β has been shown to promote cancer progression by shaping the architecture of the tumor and by suppressing the anti-tumor activities of the immune cells, thus generating an immunosuppressive environment that prevents or attenuates the efficacy of anticancer immunotherapies (44). However, no difference was found in our analysis of the immune-related signaling pathways. Such research may require further RNA expression results.
We note that several therapies targeting the TGF-β pathway are already in clinical development, and we suggest that patients with metastatic CRC actively participate in such treatments to assess the efficacy of these novel targeted therapies in combination with anti-EGFR therapy (45). TGF-β1 as the relevant isoform is emerging as a promising target for cancer therapy. The blockade of TGFβ1 in combination with other immunotherapies such as cancer vaccines increased the efficacy of a prophylactic cellular vaccine against the transplanted colon cancer model CT26 (a preclinical model) (46). Galunisertib (LY2157299) and Vactosertib (TEW-7197) as TβRI kinase inhibitors have been involved in the phase I/II trial in patients with metastatic CRC (45). Two microsatellite-stable CRC patients who received NIS793 (previously XPA-42-068), a pan anti-TGF—neutralizing antibody, achieved a partial response (PR). In preclinical mouse tumor models of CRC, bintrafusp alfa (formerly GSK-4045154, M7824, and MSB0011359C), a first-in-class investigational bifunctional fusion protein intended to block TGF-β and PD-L1, showed greater antitumor activity versus anti-PD-L1 or anti-TGF-treatment alone. TGF-β inhibitors have a number of toxicities; the most frequent treatment-related adverse events were bleeding events and TGF-β inhibition-mediated skin adverse events, but even then the future of combined targeting of the PD-1/PD-L1 pathway and TGF-β seems to be bright (47).
A previous study revealed that clusters of transverse colon tumors were more similar to left-sided tumors than right-sided tumors (20). Clustering based on the molecular profiles used to explore the distinctive molecular profiles of the patients who ignored the tumor locations. There were no differences in the three locations among the different clusters, which is consistent with the results showing gradual changes in the molecules along the bowel (19,20). We studied as large a population as possible; however, the number of patients with different tumor locations in our cohort was still small. The lack of survival information limited the further exploration of the distinctive molecular profiles of 3 locations. However, our study proposed distinctive molecular profiles for the sigmoid colon, rectosigmoid junction, and rectum, which may contribute to the selection of individualized treatment for tumors at different locations.
In this study, we identified the unique molecular features of rectosigmoid junction cancer through comparing the molecular features between rectosigmoid junction and rectum or sigmoid colon cancer. These molecular features may have clinical implications for a precision approach in the therapy, and the exploration of molecular features could be useful for discovery of potential intervention targets. Our study may contribute to further findings and research in the area of rectosigmoid junction cancer in epidemiological studies through understanding the unique molecular features.
Conclusions
We showed the characterized molecular profiles of the sigmoid colon, rectosigmoid junction, and rectum. We also observed a gradual change in the key genes of CRC along the bowel and higher TGF-β pathway alterations in the rectosigmoid junction, and rectum. Our results may contribute to the selection of individualized treatment for tumors at different locations.
Acknowledgments
We would like to thank the patients who participated in our study and their family members. We would also like to thank the staff members at all of the centers who provided assistance in this study.
Funding: This study was supported by the National Natural Science Foundation of China (grant No. 82274269), the Precision Medicine Special project of Wuxi Municipal Health Commission (No. J201801), the Wuxi Administration of Traditional Chinese Medicine Scientific Research Project (No. ZYKJ201906), and the Top Talent Support Program for Young and Middle-Aged People of Wuxi Health Committee.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-120/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-120/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-120/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-120/coif). DG and YH report that they are employed by Geneplus-Beijing Ltd., in which they performed DNA sequencing on the Gene + Seq-2000 sequencing system. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Wuxi Hospital Affiliated to Nanjing University of Chinese Medicine (No. 201809001J01-01), and each patient provided informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bae JM, Kim JH, Cho NY, et al. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer 2013;109:1004-12. [Crossref] [PubMed]
- Lee YC, Lee YL, Chuang JP, et al. Differences in survival between colon and rectal cancer from SEER data. PLoS One 2013;8:e78709. [Crossref] [PubMed]
- DʼSouza N. Definition of the Rectum: An International, Expert-based Delphi Consensus. Ann Surg 2019;270:955-9. [Crossref] [PubMed]
- Yamamoto S, Watanabe M, Hasegawa H, et al. Prospective evaluation of laparoscopic surgery for rectosigmoidal and rectal carcinoma. Dis Colon Rectum 2002;45:1648-54. [Crossref] [PubMed]
- Käser SA, Froelicher J, Li Q, et al. Adenocarcinomas of the upper third of the rectum and the rectosigmoid junction seem to have similar prognosis as colon cancers even without radiotherapy, SAKK 40/87. Langenbecks Arch Surg 2015;400:675-82. [Crossref] [PubMed]
- Suttie SA, Shaikh I, Mullen R, et al. Outcome of right- and left-sided colonic and rectal cancer following surgical resection. Colorectal Dis 2011;13:884-9. [Crossref] [PubMed]
- Guan X, Jiang Z, Ma T, et al. Radiotherapy dose led to a substantial prolongation of survival in patients with locally advanced rectosigmoid junction cancer: a large population based study. Oncotarget 2016;7:28408-19. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145-64. [Crossref] [PubMed]
- Holch JW, Heinemann V. Response to letter entitled: Distinct metastatic patterns in colorectal cancer patients. Eur J Cancer 2017;77:1-2. [Crossref] [PubMed]
- Petrelli F, Tomasello G, Borgonovo K, et al. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA Oncol 2017;3:211-9. [Crossref] [PubMed]
- Tsilimigras DI, Brodt P, Clavien PA, et al. Liver metastases. Nat Rev Dis Primers 2021;7:27. [Crossref] [PubMed]
- Park IJ, Choi GS, Lim KH, et al. Different patterns of lymphatic spread of sigmoid, rectosigmoid, and rectal cancers. Ann Surg Oncol 2008;15:3478-83. [Crossref] [PubMed]
- Kim MS, Noh JJ, Lee YY. En bloc pelvic resection of ovarian cancer with rectosigmoid colectomy: a literature review. Gland Surg 2021;10:1195-206. [Crossref] [PubMed]
- Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol 2017;28:1713-29. [Crossref] [PubMed]
- Loree JM, Kopetz S. Recent developments in the treatment of metastatic colorectal cancer. Ther Adv Med Oncol 2017;9:551-64. [Crossref] [PubMed]
- Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 2011;60:397-411. [Crossref] [PubMed]
- Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut 2011;60:116-29. [Crossref] [PubMed]
- Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 2012;61:847-54. [Crossref] [PubMed]
- Loree JM, Pereira AAL, Lam M, et al. Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes. Clin Cancer Res 2018;24:1062-72. [Crossref] [PubMed]
- Zhang Q, Feng Z, Shi S, et al. Comprehensive analysis of lncRNA-associated ceRNA network reveals the novel potential of lncRNA, miRNA and mRNA biomarkers in human rectosigmoid junction cancer. Oncol Lett 2021;21:144. [Crossref] [PubMed]
- Vieira LM, Jorge NAN, de Sousa JB, et al. Competing Endogenous RNA in Colorectal Cancer: An Analysis for Colon, Rectum, and Rectosigmoid Junction. Front Oncol 2021;11:681579. [Crossref] [PubMed]
- He Y, Wang X. Identification of molecular features correlating with tumor immunity in gastric cancer by multi-omics data analysis. Ann Transl Med 2020;8:1050. [Crossref] [PubMed]
- Lu C, Dong XR, Zhao J, et al. Association of genetic and immuno-characteristics with clinical outcomes in patients with RET-rearranged non-small cell lung cancer: a retrospective multicenter study. J Hematol Oncol 2020;13:37. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-337.e10. [Crossref] [PubMed]
- Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544-8. [Crossref] [PubMed]
- Bagati A, Kumar S, Jiang P, et al. Integrin αvβ6-TGFβ-SOX4 Pathway Drives Immune Evasion in Triple-Negative Breast Cancer. Cancer Cell 2021;39:54-67.e9. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Koboldt DC, Steinberg KM, Larson DE, et al. The next-generation sequencing revolution and its impact on genomics. Cell 2013;155:27-38. [Crossref] [PubMed]
- Nordholm-Carstensen A, Krarup PM, Jorgensen LN, et al. Occurrence and survival of synchronous pulmonary metastases in colorectal cancer: a nationwide cohort study. Eur J Cancer 2014;50:447-56. [Crossref] [PubMed]
- Mitry E, Guiu B, Cosconea S, et al. Epidemiology, management and prognosis of colorectal cancer with lung metastases: a 30-year population-based study. Gut 2010;59:1383-8. [Crossref] [PubMed]
- Santarius T, Shipley J, Brewer D, et al. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer 2010;10:59-64. [Crossref] [PubMed]
- Falch C, Mueller S, Braun M, et al. Oncological outcome of carcinomas in the rectosigmoid junction compared to the upper rectum or sigmoid colon - A retrospective cohort study. Eur J Surg Oncol 2019;45:2037-44. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018;33:125-136.e3. [Crossref] [PubMed]
- Huang YH, Lin PC, Su WC, et al. Association between Altered Oncogenic Signaling Pathways and Overall Survival of Patients with Metastatic Colorectal Cancer. Diagnostics (Basel) 2021;11:2308. [Crossref] [PubMed]
- Chen XL, Chen ZQ, Zhu SL, et al. Prognostic value of transforming growth factor-beta in patients with colorectal cancer who undergo surgery: a meta-analysis. BMC Cancer 2017;17:240. [Crossref] [PubMed]
- Itatani Y, Kawada K, Sakai Y. Transforming Growth Factor-β Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int J Mol Sci 2019;20:5822. [Crossref] [PubMed]
- Lin Z, Zhang L, Zhou J, et al. Silencing Smad4 attenuates sensitivity of colorectal cancer cells to cetuximab by promoting epithelial-mesenchymal transition. Mol Med Rep 2019;20:3735-45. [Crossref] [PubMed]
- Bedi A, Chang X, Noonan K, et al. Inhibition of TGF-β enhances the in vivo antitumor efficacy of EGF receptor-targeted therapy. Mol Cancer Ther 2012;11:2429-39. [Crossref] [PubMed]
- Dews M, Tan GS, Hultine S, et al. Masking epistasis between MYC and TGF-β pathways in antiangiogenesis-mediated colon cancer suppression. J Natl Cancer Inst 2014;106:dju043. [Crossref] [PubMed]
- Derynck R, Turley SJ, Akhurst RJ. TGFβ biology in cancer progression and immunotherapy. Nat Rev Clin Oncol 2021;18:9-34. [Crossref] [PubMed]
- Huang CY, Chung CL, Hu TH, et al. Recent progress in TGF-β inhibitors for cancer therapy. Biomed Pharmacother 2021;134:111046. [Crossref] [PubMed]
- Canè S, Van Snick J, Uyttenhove C, et al. TGFβ1 neutralization displays therapeutic efficacy through both an immunomodulatory and a non-immune tumor-intrinsic mechanism. J Immunother Cancer 2021;9:e001798. [Crossref] [PubMed]
- Lan Y, Yeung TL, Huang H, et al. Colocalized targeting of TGF-β and PD-L1 by bintrafusp alfa elicits distinct antitumor responses. J Immunother Cancer 2022;10:e004122. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


