
Function of hsa_circ_0006646 as a competing endogenous RNA to promote progression in gastric cancer by regulating the miR-665–HMGB1 axis
Highlight box
Key findings
• It has been definitely confirmed that hsa_circ_0006646 promotes the progression of GC by regulating the miR-665–HMGB1 axis.
What is known and what is new?
• miR-665 is a tumor-suppressive miRNA in GC and HMGB1 has been reported to be involved in the progression of GC.
• hsa_circ_0006646 promotes the progression of GC by upregulating HMGB1 and activating the Wnt/β-catenin signaling pathway through sponging miR-665.
What is the implication, and what should change now?
• The study provides a novel insight into the mechanism underlying the circRNA-induced progression of GC and could lead to the development of a potential biomarker and therapeutic target for treating GC.
Introduction
Gastric cancer (GC) is the fifth most common cancer and the third major cause of cancer-related death worldwide, especially in developing countries (1). Although many advances have been achieved in terms of the diagnosis and treatment of GC, the overall 5-year survival rate is still less than 30%, as most patients with GC are found to be in the advanced stage when they are first diagnosed (2). Moreover, the functions and regulatory mechanisms of precise molecules involved in GC are still little known, which has made progress in the clinical treatment of GC considerably difficult. Thus, elucidating the underlying exact mechanisms in tumorigenesis and progression of GC is essential to providing evidence for discovering novel biomarkers and prognostic indicators for early diagnosis and treatment.
Circular RNAs (circRNAs) constitute a class of noncoding RNAs characterized by a covalently closed single-stranded loop and a canonical splicing junction site (3). Emerging studies have revealed that circRNAs are involved in multiple biological processes, such as inflammation, renal fibrosis, and tumorigenesis (4,5). CircRNAs exhibit multiple functions in cellular processes including cell cycle regulation, proliferation, migration, invasion, apoptosis, and autophagy (6). Since circRNAs are stable, conserved, and abundant in cancers, some specific circRNAs have been deemed to be of great value for treating cancer patients (3). circRNAs regulate biological behavior through multiple mechanisms, with circRNA’s acting as a competing endogenous RNA (ceRNA) to sponge microRNA (miRNA) being the most common and well-studied mechanism in cancer (7). In brief, circRNAs competitively sponge miRNAs to abolish their inhibition on target messenger RNAs (mRNAs), and thereby upregulate the expression of corresponding genes (8). For instance, circUBR5 acts as a ceRNA for miR-1179 to up-regulate UBR5 and to promote malignancy of triple-negative breast cancer (9). Additionally, circFOXM1 acts as a ceRNA to upregulate SMAD2 and promote the progression of nasopharyngeal carcinoma (10). Moreover, circIFT80 functions as a ceRNA for miR-142, miR-568, and miR-634 and Promotes the Progression of Colorectal Cancer by Targeting β-Catenin (11). However, the function and exact mechanisms of circRNAs involved in tumorigenesis and progression of GC are largely unknown and need further exploration.
In the present study, we identified hsa_circ_0006646, which originates from exon 3 and exon 4 of the PTK2 gene, as a tumor-promoting circRNA in GC. hsa_circ_0006646 was upregulated in GC tissues and cell lines. High expression of hsa_circ_0006646 was positively correlated with TNM stage, lymph node invasion and poor prognosis. In addition, hsa_circ_0006646 silencing suppressed GC cell proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) in vitro. hsa_circ_0006646 knockdown inhibited GC growth in vivo. Moreover, we identified that hsa_circ_0006646 promoted the progression of GC by sponging miR-665 to relieve its downregulation of high mobility group box 1 (HMGB1) expression. Furthermore, we demonstrated that the hsa_circ_0006646–miR-665–HMGB1 axis promoted the progression and EMT of GC cells by activating the Wnt/β-catenin signaling pathway. To our knowledge, this is the first report of the role hsa_circ_0006646–miR-665–HMGB1 axis in GC and it provides potential therapeutic targets for GC. We present this article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-240/rc).
Methods
Bioinformatic analysis
The Gene Expression Omnibus (GEO) dataset GSE163416 was downloaded from the database (http://www.ncbi.nlm.nih.gov/geo/) to screen the circRNAs involved in GC. The high throughput sequencing dates were analyzed by using the “limma” package in R software (The R Foundation for Statistical Computing). The cutoff value of fold-change was set as 2. The circRNAs with P<0.05 were regarded as the differentially expressed circRNAs between GC and normal gastric tissues.
Prediction of circRNA-targeted miRNAs
To further demonstrate the potential mechanism of hsa_circ_0006646 in GC, the interacting miRNAs of hsa_circ_0006646 were predicted with circBank (http://www.circbank.cn/) and circInteractome (https://circinteractome.nia.nih.gov/).
Predicting the target genes of miRNAs
The databases of FunRich, miRWalk, TargetScan, and miRDB (12-15) were used to predict the target mRNAs of miR-665. By overlapping the results of analysis, we found the common predicted target genes exhibiting possibly regulated by miR-665.
Clinical specimens
To establish a validation cohort, GC tissues and matched adjacent normal gastric mucosal epithelial tissues were obtained from 35 pairs of patients with GC who underwent gastroscopy biopsy in the Fourth Hospital of Hebei Medical University (Shijiazhuang, China). None of the patients enrolled in this study had received preoperative treatment, including radiotherapy, chemotherapy, or immunotherapy. Each specimen was divided into 2 pieces and handled as follows: 1 piece was fixed in 10% formaldehyde for making paraffin-embedded blocks, and the other piece was immediately frozen in liquid nitrogen for extracting RNA. All patients provided informed consent. This study was conducted in accordance with the principles of the Declaration of Helsinki (as revised in 2013). Approval was granted by the ethics committee of Fourth Hospital of Hebei Medical University (No. 2019-082).
Cell culture
The normal human gastric epithelial mucosa cell line (GES-1) and GC cell line (AGS) were purchased from BNCC Biotechnology (Henan, China). Human GC cell lines (HGC27, MKN45, and MKN74) were gifted by the Research Center of Fourth Hospital of Hebei Medical University. GES-1 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, USA) medium supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Waltham, MA, USA), while the other GC cell lines were cultured in RPMI-1640 (Gibco, USA) medium supplemented with 10% FBS. Cells were incubated at 37 ℃ in a humidified atmosphere of 5% CO2.
RNA extraction and quantitative real-time polymerase reaction
The total RNAs were extracted by using TRIzol Reagent (Solarbio, Beijing, China), and genomic DNA (gDNA) were isolated with Fast Pure DNA Isolation Kit (Vazyme Biotech CO., Nanjing, China). For detecting circRNAs and mRNAs, RNA was reversely transcribed into cDNA using a Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific). For detecting miRNAs, cDNA was synthesized with the Prime Script RT Reagent Lit (Takara Bio, Kusatsu, Japan). RNA expression was quantified by quantitative real-time polymerase chain reaction (qRT-PCR) with a SYBR Green PCR Kit (Takara Bio). Relative expression was determined using the 2-ΔΔCT method (16). GAPDH was used as internal control to normalize the expression of circRNAs and mRNAs. U6 was used as internal control to normalize the expression of miRNAs. Divergent primers were used to detect the back splice junction of circRNA, and convergent primers were used to detect linear mRNA. The primers are listed in Table 1.
Table 1
| Name | Primer | Sequence (5'-3') |
|---|---|---|
| hsa_circ_0006646 | Forward primer | ATTCCGCCTCAGTCACCT |
| Reverse primer | CTAGGTATCTGTCATATTTTCCA | |
| LinearPTK2 | Forward primer | TGGGCGGAAAGAAATCCTGC |
| Reverse primer | GGCTTGACACCCTCGTTGTA | |
| GAPDH | Forward primer | AGAAGGCTGGGGCTCATTTG |
| Reverse primer | GCAGGAGGCATTGCTGATGAT | |
| miR-665 | Forward primer | ACCAGGAGGCTGAGGC |
| Reverse primer | GAACATGTCTGCGTATCTC | |
| miR-139-3p | Forward primer | GCCCTGTTGGAGTGTCGTAT |
| Reverse primer | GTATCCAGTGCGTGTCGTGG | |
| miR-1322 | Forward primer | GATGATGCTGCTGATGCTG |
| Reverse primer | GAUGAUGCUGCUGAUGCUG | |
| U6 | Forward primer | CAAGGATGACACGCAAA |
| Reverse primer | TCAACTGGTGTCGTGG | |
| HMGB1 | Forward primer | TGCAGATGACAAGCAG CCTT |
| Reverse primer | GCTGCATCAGGCTTTCCTTT |
qRT-PCR, quantitative real-time polymerase chain reaction; U6 was used as an internal control to normalize miR-665, miR-139-3p and miR-1322 expression, and GAPDH was used as a control to normalize the expressions of hsa_circ_0006646 and HMGB1;HMGB1, high mobility group box 1.
Nucleic acid electrophoresis
The complementary DNA (cDNA) and genomic DNA (gDNA) PCR products were investigated using 2% agarose gel electrophoresis with Tris-acetate-EDTA running buffer. DNA was separated with electrophoresis at 90 V for 30 min. The DNA marker was Marker L (50-500 bp) (ZOMANBIO, Beijing, China). The bands were examined using ultraviolet irradiation.
RNase R treatment
Total RNA (2 µg) from AGS and HGC27 were incubated for 15 min at 37 ℃ with or without 3 U/mg of RNase R (Thermo Fisher Scientific, USA). Then, the resulting RNA was directly reverse-transcribed with divergent primer or convergent primer using a Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, USA) and detected by PCR assay and qRT-PCR assay.
Oligonucleotide transfection
AGS and HGC27 cells were seeded in 6-well plates and cultured to 60–70% confluence before transfection. Small interfering RNAs (siRNAs) targeting the back-splicing junction of hsa_circ_0006646 (si-hsa_circ_0006646#1, si-hsa_circ_0006646#2 and si-hsa_circ_0006646#3) was designed and synthesized with GeneSeed (Guangzhou, China), while miR-665 mimics, miR-665 inhibitor, and negative control (si-NC) oligonucleotides were designed and synthesized by Biomics Biotech (Nantong, China). Lipofectamine 2000 (Invitrogen, USA) was used as transfection a medium according to the manufacturer’s protocol. Oligonucleotide sequences are listed in Table 2.
Table 2
| Definition | Sequence (5'-3') |
|---|---|
| si-hsa_circ_0006646#1 | CAGAGGAGTGGAAAATATG |
| si-hsa_circ_0006646#2 | AGAGGAGTGGAAAATATGA |
| si-hsa_circ_0006646#3 | GAGGAGTGGAAAATATGAC |
| si-NC | TTCTCCGAACGTGTCACGT |
| sh-hsa_circ_0006646#1 | CAGAGGAGTGGAAAATATGTCAAGAGCATATTTTCCACTCCTCTGTTTTTT |
| sh-hsa_circ_0006646#2 | AGAGGAGTGGAAAATATGATCAAGAGTCATATTTTCCACTCCTCTTTTTTT |
| miR-665 mimic | ACCAGGAGGCUGAGGCCCCU |
| miR-665 inhibitor | ACCAGGAGGCUGAGGCCCCU |
si, small interfering; sh, short hairpin; NC, negative control.
Plasmid construction and stable transfection
The lentivirus-containing short hairpin RNA (shRNA) targeting hsa_circ_0006646 (sh-hsa_circ_0006646#1, sh-hsa_circ_0006646#2) was purchased from GeneSeed (Guangzhou, China). The sh-hsa_circ_0006646#1 and sh-hsa_circ_0006646#2 were transfected into AGS and HGC27 cells by using Lipofectamine 2000 (Invitrogen, USA). The transfection was performed according to the manufacturer’s instructions. After being posttransfected for 48 h, the cells were selected with puromycin (2 µg/mL) for 2 weeks to construct stable cell lines. The transfection efficiency was verified with qRT-PCR. Stable transfection sequences are listed in Table 2.
Cell proliferation assay
The proliferation of AGS and HGC27 cells was evaluated by conducting Cell Counting Kit-8 (CCK-8) assay. Briefly, AGS and HGC27 cells were seeded into 96-well plates (1×103 cells/well) and cultured at 37 ℃ with 5% CO2 for 0, 24, 48, and 72 h. At the indicated time points, 10 µL of CCK-8 reagent (Servicebio, Wuhan, China) was added to the culture medium. After incubation for 2 h, the absorbance of each well was measured at 450 nm using a microplate reader (BioTek Instruments, USA).
Colony formation assay
AGS and HGC27 cells were seeded into 6-well plates at a density of 800 cells/well and incubated at 37 ℃ in a humidified atmosphere of 5% CO2 for 2 weeks. The number of clones larger than 50 cells was counted, and the colony formation rate was then calculated. Following this, the cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 20 min, and stained with a 0.1% crystal violet (Kaigen, Nanjing, China) solution for another 20 min. The colonies were photographed and counted.
Transwell assay
The Transwell chamber (Jet Biofil, Guangzhou, China) was covered with Matrigel mix (Jet Biofil) for invasion assays. AGS and HGC27 cells were seeded in upper chambers with 200 µL of serum-free medium. The bottom chamber was filled with medium and 10% FBS as the AGS and HGC27 cell chemoattractant. After 24 h, the upper chambers were fixed and then stained with 0.1% crystal violet (Kaigen) for 20 min. The cells were imaged and counted in 5 different fields.
Wound healing experiment
After the AGS and HGC27 cells were confluent to 80% in the 6-well plates, the AGS and HGC27 cells were scratched with a 200-µL micropipette tip in the center of the well to prepare a scratch wound. The cells were then incubated with serum-free medium. Representative images were captured at 0, 24, and 48 h after injury. The relative migration rate at 24 and 48 h was quantified and compared with 0 h.
Fluorescence in situ hybridization (FISH)
Digoxin-labeled hsa_circ_0006646 and miR-665 probes were designed and synthesized by Servicebio (Wuhan, China). They were used for analysis of the colocalization of hsa_circ_0006646 and miR-665 in AGS and HGC27 cells. The probe sequences were as follows: hsa_circ_0006646: 5'-DIG-GGTATCTGTCATATTTTCCACTCCTCTG-DIG-3'; and miR-665: 5'-DIG-AGGGGCCTCAGCCTCCTGGT-Dig-3'. Hybridization was performed overnight with hsa_circ_0006646 and miR-665 probes according to the manufacturer’s instructions. The images were acquired on a fluorescence microscope (Nikon, Tokyo, Japan).
Western blotting
AGS and HGC27 cells were lysed in RIPA lysis buffer (Beyotime, Shanghai, China). The membranes were blocked with 5% skimmed milk and incubated overnight at 4 ℃ with the following primary antibodies. The primary antibodies were as follows: anti-HMGB1 (1:1,000; Bioss, Beijing, China), anti-β-catenin (1:1,000; Bioss), anti-Wnt3a (1:1,000; Bioss), anti-c-Myc (1:1,000; Bioss), anti-E-cadherin (1:1,000; Servicebio), anti-N-cadherin (1:1,000; Servicebio), anti-Vimentin (1:1,000; Huabio, Hangzhou, China), anti-Snail (1:1,000; Huabio) and anti-GAPDH (1:4,000; GB11002, Servicebio) at 4 ℃ overnight. Then, the prepared membranes were incubated with secondary antibody (1:10,000, Bioss) for 2 h. Finally, the blots were visualized with an enhanced chemiluminescent reagent (Biosharp, Beijing, China).
Dual-luciferase reporter assay
The wild-type (WT) and mutated (MUT) hsa_circ_0006646 and the 3' untranslated region (3'-UTR) of HMGB1 mRNA were synthesized and inserted into pEZX-MT06 luciferase plasmids (Hanbio, Shanghai, China). Subsequently, AGS and HGC27 cells were seeded at 5×104 cells/well in 24-well plates and allowed to settle overnight. After co-transfection with the constructed luciferase plasmids and miR-665 mimics or negative control using Lipofectamine 2000 reagent (Invitrogen,USA) for 48 h, the relative luciferase activity was measured with the dual-luciferase reporter assay system (Promega, Madison, WI, USA).
RNA immunoprecipitation
RNA immunoprecipitation (RIP) assay was performed using a Magna RIP RNA-binding protein immunoprecipitation kit (BersinBio, Guangzhou, China) according to the manufacturer’s protocol. In brief, AGS and HGC27 cells were harvested and lysed in complete RIP lysis buffer after transfected with miR-665 mimics or negative control. Then, the cell extract was incubated with magnetic beads conjugated with anti-immunoglobin G (IgG) or anti-Ago2 antibody (BersinBio, Guangzhou, China) for 6 h at 4 ℃. The beads were washed and incubated with proteinase K to remove proteins. Finally, isolated RNA was extracted using TRIzol reagent (Solarbio, China), and the purified RNA was subjected to agarose gel electrophoresis and qRT-PCR analysis for detecting the enrichment of hsa_circ_0006646 and miR-665 in Ago2-immunoprecipitates.
Immunohistochemistry
The GC tissues were fixed with 10% formalin and embedded in paraffin before the sections were treated with the primary antibodies anti-HMGB1 and anti-β-catenin. After being incubated at 4 ℃ overnight, the sections were washed twice and subsequently incubated with horseradish peroxidase (HRP) polymer-conjugated secondary antibody at room temperature. These samples were then stained with 3,3-diaminobenzidine (DAB) solution and hematoxylin. Finally, the slides were observed through a microscope.
Mice xenograft model
All animal experiments were performed under a project license granted by the Ethics Committee of Fourth Hospital of Hebei Medical University (No. 2020-056) and in compliance with the Fourth Hospital of Hebei Medical University guidelines for the care and use of animals. A protocol was prepared before the study without registration. To investigate the hsa_circ_0006646 growth effect on GC cells in vivo, 4-week-old male BALB/C nude mice were acquired from Beijing Huafukang Biotechnology Co., LTD (Beijing, China), and 10 nude mice were randomly divided into 2 groups with 5 mice in each group. The AGS cells transfected with sh-hsa_circ_0006646#1 or sh-NC (negative control) were subcutaneously injected into right flank region of the legs (5×106 cells per mouse). Tumor size was measured from perpendicular axes and calculated using the following formula: volume = (length × width2)/2. Twenty-eight days later, mice were killed via spinal dislocation, and the xenograft tumors were dissected for tumor weight, Western blotting and Immunohistochemistry staining.
Statistical analysis
Statistical analyses were performed using SPSS 25.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 9 (GraphPad Software, Inc., San Diego, CA, USA). All measurement data were presented as mean±standard deviation and were analyzed using Student’s t-test. The association between hsa_circ_0006646 expression and clinicopathological features of GC was evaluated using the Chi-squared test. The survival analysis was analyzed using the Kaplan-Meier plot. Spearman rank correlation was used to analyze the association between hsa_circ_0006646 and miR-665 or between HMGB1 and miR-665 in GC specimens. All hypothesis tests were conducted by two-sided test, and the test level was 0.05. A value of P<0.05 was considered statistically significant.
Results
hsa_circ_0006646 was found to be a circRNA exhibiting high expression in GC and positively correlated with poor prognosis
To verify the critical circRNAs that contribute to the tumorigenesis and progression of GC, we first screened the upregulated circRNAs in GC tissues by analyzing a public GEO data set GSE163416. As shown in Figure 1A, hsa_circ_0006646 exhibited the most significant upregulation, among all candidate circRNAs. We then conducted qRT-PCR to validate the results from the GEO profile. As anticipated, the expression of hsa_circ_0006646 was higher in GC tissues as compared to matched normal gastric mucosal epithelium tissues (Figure 1B). We then focused on hsa_circ_0006646 for further study. We analyzed the correlation between hsa_circ_0006646 expression and clinical features. We found that hsa_circ_0006646 expression was positively correlated with TNM stage and lymph node invasion (Table 3). Furthermore, overall survival (OS) curves was completed by using the Kaplan-Meier analysis. Patients who had high levels of hsa_circ_0006646 had significantly lower overall survival rate (Figure 1C). Meanwhile, consistent with the results of GC specimens, the expression of hsa_circ_0006646 was significantly higher in GC cell lines as compared to GES-1 cells (Figure 1D). To verify whether hsa_circ_0006646 was present in GC in the form of circRNA, we investigated the structure of hsa_circ_0006646 based on circBase database annotation. The results indicated that hsa_circ_0006646 was located at chromosome 8q24.3 (NM_001199649) and derived from exon 3 and exon 4 of the PTK2 gene with a length of 394 nt (Figure 1E). Sanger sequencing confirmed that the PCR products amplified using the divergent primers contained the head-to-tail splicing site of hsa_circ_0006646, and the sequence was consistent with that of the circBase database annotation (Figure 1E). To detect whether the head-to-tail splicing of hsa_circ_0006646 results from trans-splicing or genomic rearrangements, we designed convergent primers and divergent primers, which were used to amplify PTK2 mRNA and hsa_circ_0006646, respectively. The gel electrophoresis results showed that hsa_circ_0006646 could be detected in cDNA but not gDNA, indicating that the loop structure of hsa_circ_0006646 was derived from reverse splicing (Figure 1F). Considering that circRNAs are more stable than are linear RNAs (17), we further analyzed the stability of hsa_circ_0006646 by conducting RNase R digestion assay. The results demonstrated that hsa_circ_0006646 was resistant to RNase R treatment, while linear PTK2 was significantly degraded after RNase R treatment, further confirming the circular form of hsa_circ_0006646 (Figure 1G). Taken together, these results demonstrated that hsa_circ_0006646 was an abundant, circular and stable transcript in GC.
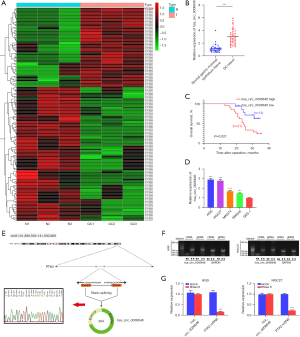
Table 3
| Characteristics | Case | hsa_circ_0006646 | χ2 | P | |
|---|---|---|---|---|---|
| High (%) | Low (%) | ||||
| Age, years | 0.326 | 0.724 | |||
| <60 | 22 | 14 (63.6) | 8 (36.4) | ||
| ≥60 | 13 | 7 (53.8) | 6 (46.2) | ||
| Gender | 0.761 | 0.477 | |||
| Male | 23 | 15 (65.2) | 8 (34.8) | ||
| Female | 12 | 6 (50.0) | 6 (50.0) | ||
| TNM stage | 9.333 | 0.006** | |||
| I–II | 10 | 2 (20.0) | 8 (80.0) | ||
| III–IV | 25 | 19 (76.0) | 6 (24.0) | ||
| Lymph node invasion | 7.619 | 0.010* | |||
| Negative (N0) | 7 | 1 (14.3) | 6 (85.7) | ||
| Positive (N1–N3) | 28 | 20 (71.4) | 8 (28.6) | ||
| Histological grade | 1.414 | 0.283 | |||
| Low | 24 | 16 (66.7) | 8 (33.3) | ||
| Middle-High | 11 | 5 (45.5) | 6 (54.5) | ||
Data were analyzed by chi-squared test. *P<0.05; **P<0.01. GC, gastric cancer; TNM, tumor-node-metastasis.
hsa_circ_0006646 knockdown suppressed the proliferation, migration, invasion, and EMT in GC cells in vitro
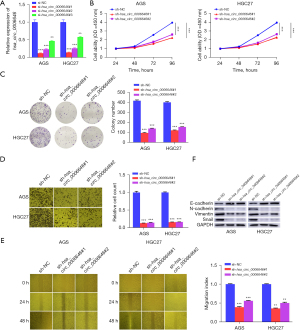
To further elucidate the functional mechanism of hsa_circ_0006646 in GC, we next investigated its effects on the malignant behaviors of GC cells. As AGS and HGC27 cell lines exhibited a relatively higher expression of hsa_circ_0006646 as compared to the other GC cell lines (Figure 1D), we performed the following experiments using these two cell lines. si-hsa_circ_0006646#1 and si-hsa_circ_0006646#2 significantly reduced the expression of hsa_circ_0006646 (Figure 2A). Then, we constructed sh-hsa_circ_0006646#1 and sh-hsa_circ_0006646#2 based on the sequences of si-hsa_circ_0006646#1 and si-hsa_circ_0006646#2 to further assess the effect of hsa_circ_0006646 on the proliferation, migration, and invasion in GC cells. Through CCK-8 assay, we found that hsa_circ_0006646 knockdown significantly suppressed the proliferation in AGS and HGC27 cells (Figure 2B). Additionally, we conducted a colony formation assay and found that the colony numbers were decreased by hsa_circ_0006646 knockdown in AGS and HGC27 cells (Figure 2C). These experiments suggested that hsa_circ_0006646 knockdown inhibited the proliferation of GC cells. Following this, wound healing and transwell assays were carried out to examine the effects of hsa_circ_0006646 on the migration and invasion of GC cells. The results demonstrated that the migration and invasion were remarkably suppressed by hsa_circ_0006646 knockdown in AGS and HGC27 cells (Figure 2D,2E). As EMT is a critical process facilitating the initiation and progression of GC, we then investigated whether EMT was involved in the tumor-promoting functions of hsa_circ_0006646. As Figure 2F shows, hsa_circ_0006646 knockdown decreased the expression of N-cadherin, Vimentin, and Snail and increased the expression of E-cadherin in AGS and HGC27 cells. Taken together, these findings suggested that hsa_circ_0006646 promoted the malignant behaviors and EMT in GC cells in vitro.
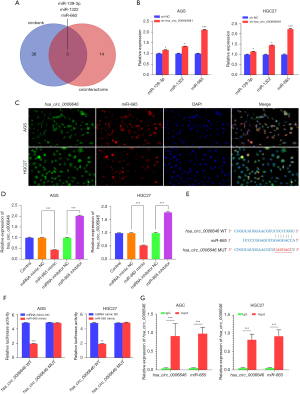
hsa_circ_0006646 bound to miR-665 in GC cells
Accumulating studies have shown that circRNAs mainly exhibited their functions in tumor progression by sponging to miRNAs and then regulating subsequent gene expression (18,19). Thus, the online bioinformatics algorithms circBank (http://www.circbank.cn/) and circInteractome (https://circinteractome.nia.nih.gov/) were adopted to predict potential miRNAs which might be sponged by hsa_circ_0006646. By overlapping the results from the above 2 algorithms, we found 3 miRNAs (miR-139-3p, miR-1322, and miR-665) as the candidates showing potential to be sponged by hsa_circ_0006646 (Figure 3A). Then, we detected their expressions in AGS and HGC27 cells after transfection of sh-hsa_circ_0006646#1 or sh-NC. The results showed that the expressions of miR-139-3p, miR-1322, and miR-665 were significantly increased in the AGS and HGC27 cells transfected with sh-hsa_circ_0006646#1 compared with the cells transfected with sh-NC (Figure 3B). As multiple studies revealed that miR-665 was a tumor-suppressive miRNA in GC, we selected miR-665 as the downstream target of hsa_circ_0006646 in subsequent study (20-22). Then, we explored the colocalization of has_circ_0006646 and miR-665 in GC cells by conducting FISH. The result demonstrated that hsa_circ_0006646 and miR-665 were mainly located in the cytoplasm of AGS and HGC27 cells (Figure 3C), suggesting that hsa_circ_0006646 had potential to sponge miR-665 in GC. To further study the correlation between miR-665 and hsa_circ_0006646, we transfected miR-665 inhibitor and miR-665 mimic into AGS and HGC27 cells and then detected the expression of hsa_circ_0006646 with qRT-PCR. As Figure 3D shows, miR-665 inhibitor and miR-665 mimic increased and decreased the expression of hsa_circ_0006646, respectively, in AGS and HGC27 cells. As there was a potential binding site on the transcript of hsa_circ_0006646 to interact with miR-665 (Figure 3E), we constructed hsa_circ_0006646-WT and hsa_circ_0006646-MUT plasmids to conduct a luciferase reporter assay. Then, we transfected hsa_circ_0006646-WT or hsa_circ_0006646-MUT plasmid into AGS and HGC27 cells and then co-transfected them with miR-665 mimics. The results showed that transfection of miR-665 mimics significantly reduced the luciferase activity in the GC cells transfected with hsa_circ_0006646-WT but did not reduce the luciferase activity of the GC cells transfected with hsa_circ_0006646-MUT, indicating that hsa_circ_0006646 acted as a ceRNA to sponge miR-665 specifically at the seed sequence (Figure 3F). Meanwhile, to determine whether hsa_circ_0006646 and miR-665 formed an RNA-induced silencing complex (RISC), an Ago2-RIP assay was conduct in AGS and HGC27 cells. As shown in Figure 3G, hsa_circ_0006646 and miR-665 were substantially enriched in Ago2-containing beads compared with those harboring control IgG, indicating that endogenous binding might occur between hsa_circ_0006646 and miR-665. In summary, hsa_circ_0006646 acted as a ceRNA to sponge miR-665 in GC cells.
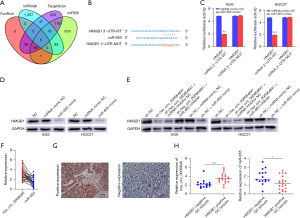
hsa_circ_0006646 upregulated HMGB1 by sponging miR-665 in GC
miRNAs interact with the 3'- UTR of target genes to regulate their expressions (23). Therefore, we predicted the downstream target of miR-665 by analyzing the bioinformatics algorithms FunRich, miRWalk, TargetScan, and miRDB (12-15). By overlapping the results of analysis, we found 6 genes exhibiting possibly regulated by miR-665: HMGB1, limb and CNS expressed 1 like (LIX1L), calcium regulated heat stable protein 1 (CARHSP1), proteasome inhibitor subunit 1 (PSMF1), solute carrier family 37 member 3 (SLC37A3), and synaptotagmin 2 (SYT2) (Figure 4A). Among these genes, only HMGB1 was reported to be involved in the progression of GC (24,25). Thus, we chose HMGB1 as the target gene of miR-665 in subsequent study. HMGB1 mRNA contains the miRNA recognition elements (MRE) of miR-665, implying that miR-665 might be the direct target of HMGB1 (Figure 4B). Furthermore, luciferase reporter plasmids with the wild type HMGB1 mRNA 3'-UTR-WT and HMGB1 mRNA 3'-UTR-MUT in the binding sites of miR-665 were constructed. The plasmids harboring HMGB1 mRNA 3'-UTR-WT or HMGB1 mRNA 3'-UTR-MUT were co-transfected into AGS and HGC27 cells with miR-665 mimic. Luciferase reporter assays showed that the miR-665 mimic significantly decreased the luciferase activity of AGS and HGC27 cells transfected with HMGB1-3'-UTR-WT, but not in those with HMGB1-3'-UTR-MUT (Figure 4C). To further validate whether HMGB1 is regulated by miR-665 in GC cells, we assayed the expression of HMGB1 in GC cells after transfection of miR-665 mimic via Western blotting. Transfection of miR-665 mimic significantly decreased the expression of HMGB1 in AGS and HGC27 cells (Figure 4D). To verify whether hsa_circ_0006646 regulates the expression of HMGB1 by sponging miR-665, we conducted a miRNA rescue experiment. As shown in Figure 4E, knockdown of hsa_circ_0006646 decreased the expression of HMGB1 in AGS and HGC27 cells while co-transfection of miR-665 inhibitor rescued this suppression. Similarly, co-transfection of miR-665 mimic downregulated the expression of HMGB1 in the AGS and HGC27 cells that were transfected with sh-NC. Next, we evaluated the hsa_circ_0006646–miR-665–HMGB1 axis in GC tissues. As anticipated, the expressions of hsa_circ_0006646 and miR-665 were negatively correlated (r=–0.3805; P=0.02; Figure 4F). Subsequently, we detected the expression of HMGB1 in GC specimens through immunohistochemistry (IHC). As shown in Figure 4G, the staining of HMGB1 was present in the nucleus of cancer cells in GC tissues. In the validation cohort, 20 cases (57.14%) exhibited a positive expression of HMGB1. Moreover, the expression of hsa_circ_0006646 was higher and the expression of miR-665 was lower in the HMGB1-positive group as compared to the HMGB1-negative group (Figure 4H). Taken together, these results provided evidence for the presence of a hsa_circ_0006646–miR-665–HMGB1 axis in GC.
hsa_circ_0006646–miR-665–HMGB1 axis promoted the proliferation, migration, invasion, and EMT in GC cells
Numerous studies have revealed that HMGB1 act as a tumor-promoter by activating the Wnt/β-catenin signaling pathway (26,27); thus, we conducted miRNA rescue experiments to clarify the effect of hsa_circ_0006646–miR-665–HMGB1 axis in GC. Western blotting assays demonstrated that knockdown of hsa_circ_0006646 in AGS and HGC27 cells decreased the expression of β-catenin, Wnt3a, and c-Myc, the key elements of Wnt/β-catenin signaling pathway, while co-transfection of miR-665 inhibitor rescued this phenomenon (Figure 5A). Consequently, knockdown of hsa_circ_0006646 decreased the expression of N-cadherin, Vimentin, and Snail and increased the expression of E-cadherin in AGS and HGC27 cells. Similarly, the above phenomenon was neutralized through co-transfection of miR-665 inhibitor (Figure 5A). We then conducted CCK-8, colony formation, wound healing, and transwell assays to determine the effects of the hsa_circ_0006646–miR-665–HMGB1 axis on the malignant biological behaviors of GC cells. The results demonstrated that the has_circ_0006646–miR-665–HMGB1 axis promoted the proliferation, colony formation, migration, and invasion in AGS and HGC27 cells (Figure 5B-5F). In summary, these data demonstrated that the hsa_circ_0006646–miR-665–HMGB1 axis promoted the progression of GC.
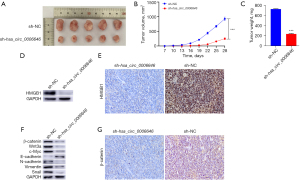
hsa_circ_0006646 enhanced the progression of GC in vivo
To further explore the effects of hsa_circ_0006646 on the progression of GC in vivo, a xenograft mouse model was established. The AGS cells transfected with sh-hsa_circ_0006646#1 or control group were subcutaneously injected into BALB/c nude mice. From the seventh day after injection, the tumor volumes were measured every 3 days to evaluate the growth of GC cells in vivo. After 28 days, the mice were killed, and the xenograft tumors were harvested and weighed. The results showed that knockdown of hsa_circ_0006646 significantly retarded the tumor growth of AGS cells in vivo (Figure 6A,6B). As shown in Figure 6C, the tumor weights of the hsa_circ_0006646 knockdown group were lighter than those of the control group. Then, we conducted Western blotting and IHC to confirm the effect of hsa_circ_0006646 knockdown on the expression of HMGB1 in vivo. As anticipated, the expression of HMGB1 was significantly downregulated in the hsa_circ_0006646 knockdown group (Figure 6D,6E). Next, we performed Western blotting to evaluate the effect of hsa_circ_0006646 knockdown on the expression of Wnt/β-catenin pathway and EMT-related proteins in vivo. This revealed that in the hsa_circ_0006646 knockdown group, the expressions of Wnt3a, β-catenin, c-Myc, N-cadherin, Vimentin, Snail were significantly decreased while the expression of E-cadherin was increased (Figure 6F). Furthermore, we detected the expression of β-catenin by performing IHC to verify the effect of the hsa_circ_0006646–miR-665–HMGB1 axis in vivo. As shown in Figure 6G, the expression of β-catenin was significantly decreased in the hsa_circ_0006646 knockdown group. Taken together, these findings demonstrated that hsa_circ_0006646 promoted the progression of GC by upregulating HMGB1 and activating the Wnt/β-catenin signaling pathway in vivo.
Discussion
circRNAs are a new series of highly stable and abundant endogenous noncoding RNA (3). CircRNAs are pervasive but heterogeneous among multiple tissues and cell lines, the generation of circRNAs is regulated by non-canonical splicing or back-splicing events (17). Previous several pieces of studies considered circRNAs as transcriptional noise and aberrant splicing by-products (28-30). However, due to the development of high-throughput sequencing and bioinformatics analysis, an increasing number of circRNAs have given rise to a new perspective on understanding the key regulators in the development and progression of cancers (31-33). Numerous circRNAs have been proven to be aberrantly upregulated in GC, and it is relatively difficult for RNase R to degrade circRNAs as they exhibit greater stability than linear RNAs in body fluids. Therefore, circRNAs show high potential value in clinical diagnosis and evaluation of prognosis (34). For instance, circRNA circSLIT2 is a novel diagnostic and prognostic biomarker for gastric cancer (35). In addition, circRNA-ABCB10 promotes gastric cancer progression by sponging miR-1915-3p to upregulate RaC1 (36). Morever, circFCHO2 promotes gastric cancer progression by activating the JAK1/STAT3 pathway via sponging miR-194-5p (37). However, only some functions of circRNAs have been clarified in GC, limiting further study on improving those treatment strategies targeting key circRNAs. Therefore, it is necessary to elucidate the functions and clinical significance of novel circRNAs involved in the progression of GC.
In this study, by analyzing the public GEO database and bioinformatics algorithms, we first identified hsa_circ_0006646 as a critical circRNA upregulated in GC. hsa_circ_0006646 was found to originate from exon 3 and exon 4 of its parent gene, PTK2. The roles of different circRNAs originating from the PTK2 gene in cancers have already been examined. For instance, hsa_circ_0005273, which originates from of exons 27, 28, and 29 of PTK2, promotes growth and metastasis of colorectal cancer (38). Moreover, circPTK2 was found to modulate migration and invasion via miR-136/NFIB signaling in triple-negative breast cancer cells in vitro (39). Regarding GC, circPTK2 suppresses the progression of GC by targeting the miR-196a-3p–AATK axis (40). Besides, circPTK2 inhibits the tumorigenesis and metastasis of GC by sponging miR-134-5p and activating CELF2/PTEN signaling (41). To our knowledge, the expression and function of hsa_circ_0006646 in GC have not been elucidated. By analyzing the GC tissues and matched normal gastric mucosal epithelium tissues, we found that hsa_circ_0006646 was significantly upregulated in GC tissues and its high expression was correlated with TNM stage, lymph node metastasis and poor prognosis of patients with GC, indicating that hsa_circ_0006646 might be a potential tumor-promoting circRNA. To reveal the biological functions of hsa_circ_0006646, we knocked down hsa_circ_0006646 expression and then evaluated its effects on the malignant behaviors in GC cells. hsa_circ_0006646 knockdown significantly inhibited the proliferation, colony formation, migration, and invasion in GC cells. Furthermore, hsa_circ_0006646 knockdown increased the expression of E-cadherin and decreased the expression of N-cadherin, Vimentin, and Snail in GC cells. Taken together, our results suggest that hsa_circ_0006646 functions as a tumor-promoting circRNA by facilitating the proliferation, colony formation, migration, invasion, and EMT in GC cells.
It has been reported that exon–intron and intron-derived circRNAs mainly function in the cell nucleus by promoting the transcription of their parent gene. In contrast, exon-derived circRNAs do not affect the expression of their parent genes due to their cytoplasmic location (42). The most well-studied functional mechanisms of cytoplasmic circRNAs include their acting as ceRNAs to sponge miRNAs and thereby regulate the expression of target genes (43). Therefore, we applied the bioinformatics algorithms circBank and circInteractome to predict the miRNAs that were potentially sponged by hsa_circ_0006646. By overlapping the analyses, we screened out 3 candidate miRNAs exhibiting potential to be the downstream miRNAs of hsa_circ_0006646 in GC. Among these 3 miRNAs, miR-665 exhibited the most significant upregulation in GC cells after hsa_circ_0006646 knockdown. Next, we conducted a FISH assay and discovered that hsa_circ_0006646 and miR-665 were co-located in the cytoplasm of GC cells. Furthermore, we performed dual-luciferase reporter and RIP assay and found that hsa_circ_0006646 directly bound with miR-665 to form a RISC. Then, we employed the bioinformatics algorithms FunRich, miRWalk, TargetScan, and miRDB to determine the downstream target of miR-665. By overlapping the results, we found 6 genes exhibiting the potential to be regulated by miR-665, among which HMGB1 was particularly noteworthy due to its concrete function in GC (24,44). HMGB1 is a protein in the high mobility group box superfamily (45). As a chromatin component common in mammalian cells, HMGB1 plays a role in the pathogenesis of inflammation, trauma, immune system diseases, and malignant tumors (46). Accumulating studies have demonstrated that HMGB1 is expressed at high level in multiple malignant tumors (47-49). HMGB1 is a key regulator in EMT, promoting the invasion, metastasis, and drug resistance in cancers by regulating various signaling pathways (50,51). For instance, HMGB1 activates the Wnt/β-catenin and RAGE/NF-κB signaling pathways to promote the EMT process of lung cancer cells (26) and prostate cancer cells, (52) respectively. Additionally, HMGB1 promotes snail-mediated EMT in glioblastoma cells via promoting the degradation of GSK-3β (53). High expression of HMGB1 has also been proven to be an indicator of poor prognosis in GC patients (54). In this study, miRNA rescue experiments confirmed there to be a hsa_circ_0006646–miR-665–HMGB1 axis in GC cells, with this axis upregulating E-cadherin and downregulating N-cadherin, Vimentin, and Snail. The downregulation of E-cadherin and upregulation of N-cadherin, Vimentin, and Snail are considered to be the fundamental processes of EMT (55). Thus, these results indicated that hsa_circ_0006646 promotes EMT in GC cells by regulating the miR-665–HMGB1 axis. As it concerns the functional mechanisms, the hsa_circ_0006646–miR-665–HMGB1 axis increased the expression of crucial elements involved in Wnt/β-catenin pathways, including β-catenin, Wnt3a, and c-Myc. As a result, knocking down hsa_circ_0006646 suppressed the proliferation, colony formation, invasion, and migration in GC cells, and miR-665 inhibitor partially abolished this suppression. Furthermore, we confirmed the existence of a hsa_circ_0006646–miR-665–HMGB1 axis in GC specimens. Therefore, the identification of the hsa_circ_0006646–miR-665–HMGB1 axis expands our knowledge of the regulatory mechanism underlying GC pathogenesis and progression.
Conclusions
To our knowledge, this is the first study to thoroughly investigate the expression, regulation, and function of hsa_circ_0006646 in GC. Our findings provide evidence for the existence and biological functions of the hsa_circ_0006646–miR-665–HMGB1 axis in GC, which promoted the proliferation, colony formation, migration, and invasion in GC by regulating Wnt/β-catenin signaling. These findings lay the foundation for further clinical study on hsa_circ_0006646–miR-665–HMGB1 axis and provide a novel insight into the mechanism underlying circRNA-induced progression of GC.
Acknowledgments
We would like to thank Prof. Huixian Cui from the International Cooperation Laboratory of Stem Cell Research, Hebei Medical University, for his kind help, as well as those patients enrolled in this study.
Funding: This work was sponsored by the National Natural Science Foundation of China (grant Nos. 81871894 and 91942314) and the Natural Science Foundation of Hebei Province (grant No. H2021206070).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-240/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-240/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-240/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-240/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Fourth Hospital of Hebei Medical University (No. 2019-082), and eligible patients signed informed consent. This study was conducted in accordance with the principles of the Declaration of Helsinki (as revised in 2013). All animal experiments were performed under a project license granted by the Ethics Committee of Fourth Hospital of Hebei Medical University (No. 2020-056) and in compliance with the Fourth Hospital of Hebei Medical University guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Ilic M, Ilic I. Epidemiology of stomach cancer. World J Gastroenterol 2022;28:1187-203. [Crossref] [PubMed]
- Chen L, Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett 2021;505:49-57. [Crossref] [PubMed]
- Norwood DA, Montalvan EE, Dominguez RL, et al. Gastric Cancer: Emerging Trends in Prevention, Diagnosis, and Treatment. Gastroenterol Clin North Am 2022;51:501-18. [Crossref] [PubMed]
- Cheng X, Ai K, Yi L, et al. The mmu_circRNA_37492/hsa_circ_0012138 function as potential ceRNA to attenuate obstructive renal fibrosis. Cell Death Dis 2022;13:207. [Crossref] [PubMed]
- Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci 2018;19. [Crossref] [PubMed]
- Zhang M, Bai X, Zeng X, et al. circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta 2021;523:120-30. [Crossref] [PubMed]
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020;21:475-90. [Crossref] [PubMed]
- Gong G, She J, Fu D, et al. CircUBR5 acts as a ceRNA for miR-1179 to up-regulate UBR5 and to promote malignancy of triple-negative breast cancer. Am J Cancer Res 2022;12:2539-57. [PubMed]
- Pei S, Ma C, Chen J, et al. CircFOXM1 acts as a ceRNA to upregulate SMAD2 and promote the progression of nasopharyngeal carcinoma. Mol Genet Genomic Med 2022;10:e1914. [Crossref] [PubMed]
- Liu N, Jiang F, Chen Z, et al. circIFT80 Functions as a ceRNA for miR-142, miR-568, and miR-634 and Promotes the Progression of Colorectal Cancer by Targeting β-Catenin. Dis Markers 2022;2022:8081246. [Crossref] [PubMed]
- Fonseka P, Pathan M, Chitti SV, et al. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol 2021;433:166747. [Crossref] [PubMed]
- Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4. [Crossref] [PubMed]
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127-d31. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Zhang W, Liu H, Jiang J, et al. CircRNA circFOXK2 facilitates oncogenesis in breast cancer via IGF2BP3/miR-370 axis. Aging (Albany NY) 2021;13:18978-92. [Crossref] [PubMed]
- Hashemi N, Jamshidian A, Babaei S, et al. Reconstruction and analysis of circRNA-miRNA-mRNA network in the pathology of lung cancer. Klin Onkol 2022;35:461-72. [Crossref] [PubMed]
- Zhang M, Wang S, Yi A, et al. microRNA-665 is down-regulated in gastric cancer and inhibits proliferation, invasion, and EMT by targeting PPP2R2A. Cell Biochem Funct 2020;38:409-18. [Crossref] [PubMed]
- Fang X, Bai Y, Zhang L, et al. MicroRNA-665 regulates the proliferation, apoptosis and adhesion of gastric cancer cells by binding to cadherin 3. Oncol Lett 2021;21:494. [Crossref] [PubMed]
- He C, Liu Y, Li J, et al. LncRNA RPSAP52 promotes cell proliferation and inhibits cell apoptosis via modulating miR-665/STAT3 in gastric cancer. Bioengineered 2022;13:8699-711. [Crossref] [PubMed]
- Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer 2021;1875:188491. [Crossref] [PubMed]
- Tang T, Wang S, Cai T, et al. High mobility group box 1 regulates gastric cancer cell proliferation and migration via RAGE-mTOR/ERK feedback loop. J Cancer 2021;12:518-29. [Crossref] [PubMed]
- Takaki W, Konishi H, Matsubara D, et al. Role of Extracellular High-Mobility Group Box-1 as a Therapeutic Target of Gastric Cancer. Int J Mol Sci 2022;23. [Crossref] [PubMed]
- Wang XH, Zhang SY, Shi M, et al. HMGB1 Promotes the Proliferation and Metastasis of Lung Cancer by Activating the Wnt/β-Catenin Pathway. Technol Cancer Res Treat 2020;19:1533033820948054. [Crossref] [PubMed]
- Wang S, Du S, Lv Y, et al. MicroRNA-665 inhibits the oncogenicity of retinoblastoma by directly targeting high-mobility group box 1 and inactivating the Wnt/β-catenin pathway. Cancer Manag Res 2019;11:3111-23. [Crossref] [PubMed]
- Lee ECS, Elhassan SAM, Lim GPL, et al. The roles of circular RNAs in human development and diseases. Biomed Pharmacother 2019;111:198-208. [Crossref] [PubMed]
- Santer L, Bär C, Thum T. Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Mol Ther 2019;27:1350-63. [Crossref] [PubMed]
- Nisar S, Bhat AA, Singh M, et al. Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Front Cell Dev Biol 2021;9:617281. [Crossref] [PubMed]
- Zhang W, Liu T, Li T, et al. Hsa_circRNA_102002 facilitates metastasis of papillary thyroid cancer through regulating miR-488-3p/HAS2 axis. Cancer Gene Ther 2021;28:279-93. [Crossref] [PubMed]
- Zheng L, Liang H, Zhang Q, et al. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol Cancer 2022;21:41. [Crossref] [PubMed]
- Xi Y, Shen Y, Wu D, et al. CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol Cancer 2022;21:145. [Crossref] [PubMed]
- Ghafouri-Fard S, Honarmand Tamizkar K, Jamali E, et al. Contribution of circRNAs in gastric cancer. Pathol Res Pract 2021;227:153640. [Crossref] [PubMed]
- Wang L, Xiao S, Zheng Y, et al. CircRNA circSLIT2 is a novel diagnostic and prognostic biomarker for gastric cancer. Wien Klin Wochenschr 2023; [Crossref] [PubMed]
- Liu J, Qiu G, Wang H, et al. CircRNA-ABCB10 promotes gastric cancer progression by sponging miR-1915-3p to upregulate RaC1. Dig Liver Dis 2022;54:896-904. [Crossref] [PubMed]
- Zhang Z, Sun C, Zheng Y, et al. circFCHO2 promotes gastric cancer progression by activating the JAK1/STAT3 pathway via sponging miR-194-5p. Cell Cycle 2022;21:2145-64. [Crossref] [PubMed]
- Yang H, Li X, Meng Q, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer 2020;19:13. [Crossref] [PubMed]
- Wang M, Chen D, Zhang H, et al. Circular RNA circPTK2 modulates migration and invasion via miR-136/NFIB signaling on triple-negative breast cancer cells in vitro. Inflamm Res 2022;71:409-21. [Crossref] [PubMed]
- Gao L, Xia T, Qin M, et al. CircPTK2 Suppresses the Progression of Gastric Cancer by Targeting the MiR-196a-3p/AATK Axis. Front Oncol 2021;11:706415. [Crossref] [PubMed]
- Fan HN, Zhao XY, Liang R, et al. CircPTK2 inhibits the tumorigenesis and metastasis of gastric cancer by sponging miR-134-5p and activating CELF2/PTEN signaling. Pathol Res Pract 2021;227:153615. [Crossref] [PubMed]
- Cortés-López M, Miura P. Emerging Functions of Circular RNAs. Yale J Biol Med 2016;89:527-37. [PubMed]
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22-36. [Crossref] [PubMed]
- Chung HW, Lim JB. High-mobility group box-1 contributes tumor angiogenesis under interleukin-8 mediation during gastric cancer progression. Cancer Sci 2017;108:1594-601. [Crossref] [PubMed]
- Ye M, Hou H, Shen M, et al. Circular RNA circFOXM1 Plays a Role in Papillary Thyroid Carcinoma by Sponging miR-1179 and Regulating HMGB1 Expression. Mol Ther Nucleic Acids 2020;19:741-50. [Crossref] [PubMed]
- Xue J, Suarez JS, Minaai M, et al. HMGB1 as a therapeutic target in disease. J Cell Physiol 2021;236:3406-19. [Crossref] [PubMed]
- Wang S, Zhang Y. HMGB1 in inflammation and cancer. J Hematol Oncol 2020;13:116. [Crossref] [PubMed]
- Handke NA, Rupp ABA, Trimpop N, et al. Soluble High Mobility Group Box 1 (HMGB1) Is a Promising Biomarker for Prediction of Therapy Response and Prognosis in Advanced Lung Cancer Patients. Diagnostics (Basel) 2021;11. [Crossref] [PubMed]
- Lv DJ, Song XL, Huang B, et al. HMGB1 Promotes Prostate Cancer Development and Metastasis by Interacting with Brahma-Related Gene 1 and Activating the Akt Signaling Pathway. Theranostics 2019;9:5166-82. [Crossref] [PubMed]
- Luan X, Ma C, Wang P, et al. HMGB1 is negatively correlated with the development of endometrial carcinoma and prevents cancer cell invasion and metastasis by inhibiting the process of epithelial-to-mesenchymal transition. Onco Targets Ther 2017;10:1389-402. [Crossref] [PubMed]
- Lei X, Hu X, Zhang T, et al. HMGB1 release promotes paclitaxel resistance in castration-resistant prostate cancer cells via activating c-Myc expression. Cell Signal 2020;72:109631. [Crossref] [PubMed]
- Zhang J, Shao S, Han D, et al. High mobility group box 1 promotes the epithelial-to-mesenchymal transition in prostate cancer PC3 cells via the RAGE/NF-κB signaling pathway. Int J Oncol 2018;53:659-71. [Crossref] [PubMed]
- Li H, Li J, Zhang G, et al. HMGB1-Induced p62 Overexpression Promotes Snail-Mediated Epithelial-Mesenchymal Transition in Glioblastoma Cells via the Degradation of GSK-3β. Theranostics 2019;9:1909-22. [Crossref] [PubMed]
- Wu Z, Huang Y, Yuan W, et al. Expression, tumor immune infiltration, and prognostic impact of HMGs in gastric cancer. Front Oncol 2022;12:1056917. [Crossref] [PubMed]
- Lambert AW, Weinberg RA. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer 2021;21:325-38. [Crossref] [PubMed]
(English Language Editor: J. Gray)


