
LncASAP1-IT1 promotes hepatocellular carcinoma progression through the regulation of the miR-1294/TGFBR1 pathway in vitro and in vivo
Highlight box
Key findings
• We found that the long non-coding RNA ASAP1-IT1 regulated cell proliferation, invasion, migration, and epithelial-mesenchymal transition (EMT) progression and enhanced sensitivity to sorafenib in hepatocellular carcinoma (HCC) by targeting microRNA-1294 (miR-1294)/transforming growth factor beta receptor 1 (TGFBR1) and identified a new regulatory mechanism in HCC.
What is known and what is new?
• ASAP1-IT1 is upregulated in HCC tissues and cells.
• The knockdown of ASAP1-IT1 impeded cell proliferation, migration, invasion, and EMT progression and enhanced sensitivity to sorafenib in HCC through the miR-1294/TGFBR1 axis.
What is the implication, and what should change now?
• We identified a new biomarker and potential therapeutic target for the prognosis and treatment of HCC.
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent pathological subtype of primary liver cancer (75–85%) with a high mortality rate worldwide (1). The pathogenesis of liver cancer is commonly thought to be complicated by various genetic and environmental factors (2-4). In recent years, long non-coding RNAs (lncRNAs) have been shown to be critical in regulating a wide range of biological and pathological processes, including the formation of tumors (5). LncASAP1-IT1 has been reported to regulate the development and the progression of various types of cancer, including lung, bladder, and ovarian cancer (6-8). Fu et al. reported that lncASAP1-IT1 was highly expressed in early-stage epithelial ovarian cancer and low-grade tumors (6). Yang et al.’s findings suggest that lncASAP1-IT1 promotes the stemness of bladder cancer and is associated with a poor prognosis (7). In non-small cell lung cancer, Zhang et al. found that lncASAP1-IT1 induced cytotoxic behaviors in cancer cells, such as proliferation, invasion, and metastasis, by activating the PTEN/AKT pathway (8). LncASAP1-IT1 was reported to be upregulated in 3 different HCC cell lines in contrast to normal human hepatocytes, evidenced by searching lncRNA expression profile (GSE25859). However, the role of lncASAP1-IT1 in HCC development remains to be investigated.
LncRNAs bind with micro RNAs (miRNAs) bearing complementary sequences and serve a “sponge” function to re-activate target messenger RNA, and some lncRNAs bind with multiple miRNAs. This competitive endogenous RNA (ceRNA) pathway has been linked to various illnesses, including cancer. Our bioinformatic analysis indicated that miR-1294 may bind with lncASAP1-IT1. miR-1294 has been reported to be involved in the regulation of multiple types of cancer, such as gastric cancer, breast cancer, ovarian cancer, osteosarcoma, pancreatic ductal adenocarcinoma, and HCC (9-11). A study has reported that ceRNA networks exhibit significant potential in the diagnosis and targeted therapy of cancer. However, the function of lncASAP1-IT1/miR-1294 in HCC remains incompletely defined (12).
In the present study, we hypothesized that lncASAP1-IT1 controls the miR-1294/transforming growth factor beta receptor 1 (TGFBR1) signaling axis during the development of HCC. Thus, we investigated the possible significance of lncASAP1-IT1 in HCC and its molecular mechanism. We present this article in accordance with the ARRIVE and MDAR reporting checklists (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-327/rc).
Methods
Human liver cancer tumor samples and cell lines
In total, 30 paired human HCC and adjacent histologically normal tissues (>2 cm from the tumors’ edges) were collected from HCC patients (20 male and 10 female; age range: 40–73 years) who underwent surgical treatment at The Fourth Hospital of Hebei Medical University. During the surgery, samples were taken from patients, and the diagnosis was verified by skilled pathologists. Patients were excluded from the study if they had previously undergone radiation or chemotherapy, had a history of malignancy, or had not provided informed consent. Real-time-quantitative polymerase chain reaction (RT-qPCR) was used to determine the expression levels of lncASAP1-IT1 in the liver tissues. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Fourth Hospital of Hebei Medical University (No. 2022KS025) and informed consent was taken from all the patients.
The Shanghai Cell Bank provided the THLE-2 normal human cell line and the Bel-7405, Huh7, Hep3B, and Sk-Hep1 malignant cell lines.
Bioinformatics prediction
The interaction between miR-1294 and lncASAP1-IT1 was predicted by DIANA TOOLS (http://diana.imis.athena-innovation.gr) and LncBook (https://ngdc.cncb.ac.cn/lncbook/interaction). MiRDB (http://www.mirdb.org/), Targetscan (http://www.targetscan.org/), and Starbase (http://starbase.sysu.edu.cn/) predicted the potential target gene TGFBR1 that could be targeted by miR-1294.
Luciferase reporter assays
The Dual-Luciferase Reporter Assay System (Promega, Shanghai) was used to quantify Firefly and Renilla luciferase activity (13).
Cell Counting Kit 8 (CCK-8) assays
The cell suspensions of each group were collected, and 100 µL of cells (1×106 cells/well) were inoculated into 96-well plates; each group comprised 3 replicate wells. After 48 hours of growth, 10 µL of CCK-8 reagent (Shanghai Biyuntian Co., Ltd.) was added to each well and incubated at 37 ℃ for 1 h. The absorbance was read at 490 nm with a microplate reader (14).
Colony formation assays
100 cells were counted and seeded in triplicate in a 6-well plate, and cultured continuously for 14 days. Culture was stopped when visible clones appeared in the petri dish. After discarding the supernatant, the cells were washed with phosphate buffered saline. The cells were fixed with 4% paraformaldehyde for 15 min, stained with 0.1% crystal violet for 10 min, and air-dried. The number of effective clones with at least 15 cells was counted under low magnification (15).
Transwell assays
Transwell chambers coated (invasion) or uncoated (migration) with Matrigel (BD, USA) were placed in a 24-well cell plate, and cell solution containing 105 cells was added to the upper chamber. Dulbecco’s Modified Eagle Medium (500 µL) containing 10% fetal bovine serum was then added to the lower chamber. After 24 h of incubation, the chamber was removed. After washing the cells with phosphate buffered saline, the remaining cells on the filter membrane were carefully wiped off with a cotton swab, and 4% paraformaldehyde and 0.5% crystal violet were added for fixation and staining, respectively, for 15 min. We then randomly selected 3 visual fields under the microscope to observe the number of transmembrane cells (16).
Nude mouse tumorigenicity assays
A total of 10 female BALB/c nude mice (aged 4–6 weeks old and weighing 18–20 g) were acquired from the Vital River Laboratory Animal Center (Beijing, China). The nude mice were randomly allocated to two groups (the NC and shASAP1-IT1 groups), of 5 mice per group, based on their body weight, and reared in a specific pathogen-free environment. Next, 2×106 cells were implanted subcutaneously into the nude mice to form tumors. When the maximum diameter of the tumors reached about 1 cm, the nude mice underwent uniform euthanasia, and the tumor tissues were stripped, photographed, and weighed. Formalin-fixed and paraffin-embedded tissue sections were incubated with TGFBR1 primary antibody (dilution 1:200; Cell Signaling Technology), Ki67 antibody (dilution 1:200; Proteintech), and CD34 (dilution 1:200; Abcam) overnight at 4 ℃ and the Horseradish peroxidase (HRP)-labeled secondary antibodies (dilution 1:2,000; Abcam) were then incubated. Animal experiments were performed under a project license (No. 2019033) granted by the ethics committee of the Fourth Hospital of Hebei Medical University, in compliance with national guidelines for the care and use of animals. A protocol was prepared before the study without registration.
RT-qPCR
Complementary DNA was generated by reverse transcription using RNA as the template, according to the instructions of the PrimeScript RT Reagent Kit (Invitrogen, Thermo Fisher Scientific). RT-qPCR was performed on an ABI 7500 PCR System using SYBR-Green Master Mix (Invitrogen, Thermo Fisher Scientific). The following primer sequences were used:
- ASAP1-IT1: forward: 5'-AAACATCATCCCCAGAGTGG-3';
- ASAP1-IT1: reverse: 5'-GCCTTGCTCACCTCTGAAAC-3';
- miR-1294: forward: 5'-CTCACGAGAGAGGAAGGCA-3';
- miR-1294: reverse: 5'-ACCTCAAGAACAGTATTTCCAGG-3';
- U6: forward: 5'-CGCTTCACGAATTTGCGTGTCAT-3';
- U6: reverse: 5'-CGCTTCACGAATTTGCGTGTCAT-3';
- TGFBR1: forward: 5'-TGCCATAACCGCACTGTCA-3';
- TGFBR1: reverse: 5'-AATGAAAGGGCGATCTAGTGATG-3';
- GAPDH: forward: 5'-CGCTCTCTGCTCCTCCTGTTC-3';
- GAPDH: reverse: 5'-ATCCGTTGACTCCGACCTTCAC-3'.
Western blot
The cells were collected from each group during the logarithmic growth phase, and radio immunoprecipitation assay (RIPA) lysis buffer lysate was added for lysis and centrifugation to extract the total cell proteins. The cell protein concentration was determined using the bicinchoninic acid (BCA) method. Protein samples of 40 µg were thoroughly mixed with loading buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) then transferred on polyvinylidene fluoride (PVDF) membranes. The membranes were incubated overnight at 4 ℃ with primary antibodies against E-cadherin, N-cadherin, TGFBR1, β-actin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (all 1:1,000, from Cell Signaling Technology) were added, and at 4 ℃ overnight. The membrane was then washed with Tris-buffered saline-Tween-20 buffer (TBST), secondary antibodies (Abcam, 1:10,000) for 2 h at room temperature, and TBST washed the membrane. Protein bands were developed by a standard enhanced chemiluminescence (ECL) kit (Solarbio), and observed via a gel image analysis system (Bio-Rad).
Statistical analysis
The data were analyzed using GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA). An analysis of variance or a Student’s t-test was used. A two-sided P value <0.05 was considered statistically significant.
Results
LncASAP1-IT1 is overexpressed in liver cancer tissue and liver cancer cell lines
The expression of lncASAP1-IT1 was initially assessed by RT-qPCR to examine its involvement in liver cancer pathogenesis and pathogenesis. We discovered that lncASAP1-IT1 was significantly more highly expressed in the liver cancer tissues than the healthy liver tissues. Further, the expression level of lncASAP1-IT1 was significantly higher in the HCC lines (i.e., Bel-7405, Huh7, Hep3B, and Sk-Hep1) than the normal human cell line (i.e., THLE-2) (Figure 1).

Knocking down lncASAP1-IT1 significantly suppresses the cytotoxic behaviors of liver cancer cells
RT-qPCR revealed that compared to the control group, the transfection of sh-lncASAP1-IT1-1 and sh-lncASAP1-IT1-2 significantly decreased the level of lncASAP1-IT1-1 in the Bel-7405 and Hep-3B cells (Figure 2A). The knockdown of lncASAP1-IT1 inhibited the survival rates of the Bel-7405 and Hep-3B cells (Figure 2B). The nuclear condensation in the shASAP1-IT1 groups that underwent Giemsa staining by microscopic observation is shown in Figure 2C. A colony formation experiment was also conducted to investigate the effect of lncASAP1-IT1 on the proliferative abilities of the Bel-7405 and Hep-3B cells. Our results showed that lncASAP1-IT1 knockdown significantly decreased the colony numbers in both the Bel-7405 and Hep-3B cells (Figure 2D). The transwell assay results showed that lncASAP1-IT1 knockdown reduced Bel-7405 and Hep-3B cell migration and invasion capacities (Figure 2E,2F). These findings indicated that lncASAP1-IT1 knockdown decreased HCC cell growth and migration.

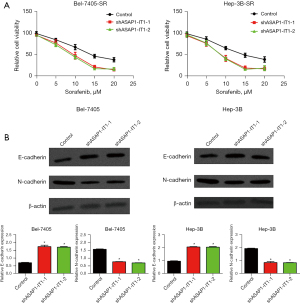
LncASAP1-IT1 contributes to HCC cell lines sorafenib resistance and ETM
To better understand lncASAP1-IT1 function in chemoresistance, we generated sorafenib-resistant HCC lines (using Bel-7405-SR Hep3B-SR) and evaluated drug resistance using the CCK-8 test. As seen in Figure 3A, we found that silencing lncASAP1-IT1 enhanced the sorafenib sensitivity of the HCC cells. We then sought to decipher how ASAP1-IT1 contributes to epithelial-mesenchymal transition (EMT). The Western blot analysis revealed that compared to the control, interfering with ASAP1-IT1 increased E-cadherin expression but decreased N-cadherin expression (Figure 3B).
LncASAP1-IT1 acts as a sponge for miR-1294
As predicted by the DIANA and LncBase tools, we found that lncASAP1-IT1 directly binds to miR-1294 (Figure 4A). We then used RNA mimics to overexpress miR-1294 and miR-1294 inhibitors to downregulate its expression in the Bel-7405 cells (Figure 4B). To establish the binding of miR-1294 to lncASAP1-IT1, we used luciferase assays. Our findings indicated that lncASAP1-IT1 binds directly to miR-1294 (Figure 4C). Conversely, silencing lncASAP1-IT1 resulted in a reduction in miR-1294 expression in comparison to the control group (Figure 4D).

TGFBR1 is the downstream target of miR-1294
The data indicated that miR-1294 might target TGFBR1 (Figure 5A). Compared to the control groups, the miR-1294 mimics significantly inhibited TGFBR1 expression, and the miR-1294 inhibitors significantly increased TGFBR1 expression (Figure 5B). These findings provided further evidence of the effect of miR-1294 on TGFBR1 expression. The regulation of miR-1294 on TGFBR1 was subsequently confirmed by a luciferase experiment. The results indicated that the miR-1294 mimic significantly reduced the luciferase activity of wild-type TGFBR1 (Figure 5C). Additionally, silencing lncASAP1-IT1 lowered TGFBR1 expression compared to the control group, but silencing miR-1294 reversed the decrease in TGFBR1 expression induced by shASAP1-IT1 (Figure 5D). In terms of cytotoxic behavior modulation, we assessed the migratory ability of the liver cancer cells using the transwell test. The overexpression of miR-1294, silencing of lncASAP1-IT1, and silencing of TGFBR1 all resulted in a substantial reduction in Bel-7405 cell movement compared to the control. Additionally, the treatment with the miR-1294 inhibitors restored the decreased migratory abilities of the Bel-7405 cells caused by the TGFBR1 knockdown, while the overexpression of TGFBR1 restored the reduced migration abilities of the Bel-7405 cells induced by the shASAP1-IT1 (Figure 5E).

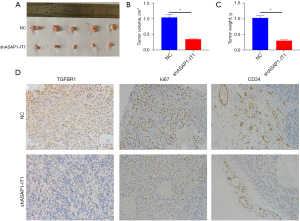
LncASAP1-IT1 suppressed the liver cancer growth of the xenograft tumors in vivo
Bel-7405 cells expressing shASAP1-IT1 or sh-NC were implanted into the subcutaneous region of the necks of the nude mice. After dissection, the tumor volume was determined, and the shASAP1-IT1 group had a larger tumor volume than the control group (Figure 6A-6C). The immunohistochemistry staining showed the lower expression of TGFBR1 and the proliferation and angiogenesis markers (Ki-67 and CD34) in the shASAP1-IT1 group compared with the NC group (Figure 6D).
Discussion
HCC is a prevalent liver cancer globally. Despite breakthroughs in clinical intervention tactics, high rates of recurrence and metastasis impede the therapeutic efficacy and result in poor outcomes (2,4,17). Thus, knowledge of the process driving carcinogenesis is required to diagnose and treat HCC early.
LncRNAs have been reported to be critical non-coding RNAs that regulate liver cancer progression in various aspects of the cellular functions of liver cancer cells, including proliferation, apoptosis, migration, and invasion (18,19). For example, Xu et al. showed that lncRNA RP11-386G11.10 was overexpressed in HCC and positively correlated with tumor size, tumor node metastasis stage, and a poor prognosis in HCC patients (20). Moreover, linc Hes family bHLH transcription factor 6 (lncSHRG) has been shown to accelerate HCC cell progression by targeting miR26a/SKP2 axis (21). The current study found that lncASAP1-IT1 expression was significantly elevated in the HCC tissues compared to the adjacent healthy tissues. Experiments on the cytotoxic behaviors of HCC cells suggest that lncASAP1-IT1 regulates the proliferation, migration, invasion, and sorafenib sensitivity of the Bel-7405 and Hep-3B cells.
In the present study, we also demonstrated that lncASAP1-IT1 regulates the target gene TGFBR1 by binding with miR-1294 by the ceRNA mechanism. Many studies have indicated that miR-1294 is a suppressor in the pathogenesis of HCC (22,23). We found that lncASAP1-IT1 regulates HCC tumorigeneses by facilitating miR-1294. TGFBR1 has been reported to be related to the development of colorectal cancer, enhance the migration and invasion of breast cancer cells and participate in bladder cancer and non-small cell lung cancer (24-27). However, further research needs to be conducted to determine if TGFBR1 regulates liver cancer cells. Our results suggest that the upregulation of TGFBR1 through miR-1294 promotes the migration abilities of liver cancer cells and thus suggests that TGFBR1 plays a role in promoting liver cancer progression. In addition, there are some problems and challenges that need to be overcome in the clinical application of lncRNA, such as delivery, specificity and tolerability.
Conclusions
Our findings suggest that lncASAP1-IT1 is highly expressed in HCC and supports its progression through the miR-1294/TGFBR1 axis. The inhibition of ASAP1-IT1 impedes cell proliferation, migration, invasion, and EMT progression, and enhances sensitivity of HCC cells to sorafenib in HCC. Taken together, our results provide new insight into the role of lncRNAs in the development of HCC and support the notion that lncASAP1-IT1 may serve as a prognostic biomarker and a potential therapeutic molecular target for the treatment of HCC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-327/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-327/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-327/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-327/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were performed under a project license (No. 2019033) granted by the ethics committee of the Fourth Hospital of Hebei Medical University, in compliance with national guidelines for the care and use of animals. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Fourth Hospital of Hebei Medical University (No. 2022KS025) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nagaraju GP, Dariya B, Kasa P, et al. Epigenetics in hepatocellular carcinoma. Semin Cancer Biol 2022;86:622-32. [Crossref] [PubMed]
- Lazăr DC, Avram MF, Romoșan I, et al. Malignant hepatic vascular tumors in adults: Characteristics, diagnostic difficulties and current management. World J Clin Oncol 2019;10:110-35. [Crossref] [PubMed]
- van Tienderen GS, Groot Koerkamp B, IJzermans JNM, et al. Recreating Tumour Complexity in a Dish: Organoid Models to Study Liver Cancer Cells and their Extracellular Environment. Cancers (Basel) 2019;11:1706. [Crossref] [PubMed]
- Turley EA, Veiseh M, Radisky DC, et al. Mechanisms of disease: epithelial-mesenchymal transition--does cellular plasticity fuel neoplastic progression? Nat Clin Pract Oncol 2008;5:280-90. [Crossref] [PubMed]
- Yang M, Lu H, Liu J, et al. lncRNAfunc: a knowledgebase of lncRNA function in human cancer. Nucleic Acids Res 2022;50:D1295-306. [Crossref] [PubMed]
- Fu Y, Biglia N, Wang Z, et al. Long non-coding RNAs, ASAP1-IT1, FAM215A, and LINC00472, in epithelial ovarian cancer. Gynecol Oncol 2016;143:642-9. [Crossref] [PubMed]
- Yang L, Xue Y, Liu J, et al. Long noncoding RNA ASAP1-IT1 promotes cancer stemness and predicts a poor prognosis in patients with bladder cancer. Neoplasma 2017;64:847-55. [Crossref] [PubMed]
- Zhang L, Shi SB, Zhu Y, et al. Long non-coding RNA ASAP1-IT1 promotes cell proliferation, invasion and metastasis through the PTEN/AKT signaling axis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2018;22:142-9. [PubMed]
- Zhang Y, Huang S, Guo Y, et al. MiR-1294 confers cisplatin resistance in ovarian Cancer cells by targeting IGF1R. Biomed Pharmacother 2018;106:1357-63. [Crossref] [PubMed]
- Wang Y, Liu G, Sun S, et al. miR-1294 alleviates epithelial-mesenchymal transition by repressing FOXK1 in gastric cancer. Genes Genomics 2020;42:217-24. [Crossref] [PubMed]
- Cai X, Yu L, Chen Z, et al. Arsenic trioxide-induced upregulation of miR-1294 suppresses tumor growth in hepatocellular carcinoma by targeting TEAD1 and PIM1. Cancer Biomark 2020;28:221-30. [Crossref] [PubMed]
- Zhu J, Wang L, Zhou Y, et al. Comprehensive analysis of the relationship between competitive endogenous RNA (ceRNA) networks and tumor infiltrating-cells in hepatocellular carcinoma. J Gastrointest Oncol 2020;11:1381-98. [Crossref] [PubMed]
- Cao JX, Lu Y, Qi JJ, et al. MiR-630 inhibits proliferation by targeting CDC7 kinase, but maintains the apoptotic balance by targeting multiple modulators in human lung cancer A549 cells. Cell Death Dis 2014;5:e1426. [Crossref] [PubMed]
- Li Y, Wu A, Chen L, et al. Hsa_circ_0000098 is a novel therapeutic target that promotes hepatocellular carcinoma development and resistance to doxorubicin. J Exp Clin Cancer Res 2022;41:267. [Crossref] [PubMed]
- Xing L, Tang X, Wu K, et al. LncRNA HAND2-AS1 suppressed the growth of triple negative breast cancer via reducing secretion of MSCs derived exosomal miR-106a-5p. Aging (Albany NY) 2020;13:424-36. [Crossref] [PubMed]
- Kong X, Zheng Z, Song G, et al. Over-Expression of GUSB Leads to Primary Resistance of Anti-PD1 Therapy in Hepatocellular Carcinoma. Front Immunol 2022;13:876048. [Crossref] [PubMed]
- Francica G, Borzio M. Status of, and strategies for improving, adherence to HCC screening and surveillance. J Hepatocell Carcinoma 2019;6:131-41. [Crossref] [PubMed]
- Huang Z, Zhou JK, Peng Y, et al. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer 2020;19:77. [Crossref] [PubMed]
- Yuan K, Lan J, Xu L, et al. Long noncoding RNA TLNC1 promotes the growth and metastasis of liver cancer via inhibition of p53 signaling. Mol Cancer 2022;21:105. [Crossref] [PubMed]
- Xu K, Xia P, Gongye X, et al. A novel lncRNA RP11-386G11.10 reprograms lipid metabolism to promote hepatocellular carcinoma progression. Mol Metab 2022;63:101540. [Crossref] [PubMed]
- Hu JJ, Zhou C, Luo X, et al. Linc-SCRG1 accelerates progression of hepatocellular carcinoma as a ceRNA of miR26a to derepress SKP2. J Exp Clin Cancer Res 2021;40:26. [Crossref] [PubMed]
- Luo Z, Lu L, Tang Q, et al. CircCAMSAP1 promotes hepatocellular carcinoma progression through miR-1294/GRAMD1A pathway. J Cell Mol Med 2021;25:3793-802. [Crossref] [PubMed]
- Lin G, Li J, Chen K, et al. Circ_0000854 regulates the progression of hepatocellular carcinoma through miR-1294 /IRGQ axis. Clin Immunol 2022;238:109007. [Crossref] [PubMed]
- Rosman DS, Phukan S, Huang CC, et al. TGFBR1*6A enhances the migration and invasion of MCF-7 breast cancer cells through RhoA activation. Cancer Res 2008;68:1319-28. [Crossref] [PubMed]
- van Tilborg AA, de Vries A, Zwarthoff EC. The chromosome 9q genes TGFBR1, TSC1, and ZNF189 are rarely mutated in bladder cancer. J Pathol 2001;194:76-80. [Crossref] [PubMed]
- Zeng Q, Phukan S, Xu Y, et al. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res 2009;69:678-86. [Crossref] [PubMed]
- Lei Z, Liu RY, Zhao J, et al. TGFBR1 haplotypes and risk of non-small-cell lung cancer. Cancer Res 2009;69:7046-52. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


