
Screening of key genes related to ferroptosis and a molecular interaction network analysis in colorectal cancer using machine learning and bioinformatics
Highlight box
Key findings
• Our study identified 4 ferroptosis-related DEGs in CRC (i.e., NOX4, TFR2, ALOXE3, and CA9), and further validated their relationship with immune cell infiltration and the associated immune checkpoints.
What is known and what is new?
• Many researchers view the molecular prognostic model of ferroptosis-related genes as a prognostic biomarker for cancer patients.
• Our findings confirm the influence of the immune microenvironment on CRC. Low NOX4 levels were more favorable to patient outcomes.
What is the implication, and what should change now?
• Our findings may facilitate future clinical diagnoses and outcome assessments of CRC.
Introduction
Colorectal cancer (CRC) is the 3rd most prevalent neoplasm worldwide (1). It is estimated that the global burden of CRC will increase to over 2.2 million new cases and 1.1 million annual deaths by the year 2030 (1). The 5-year survival rate of metastatic colon carcinoma patients is only 14% (2). In China, the incidence of CRC continues to increase, and it is arguably one of the most prevalent malignancies after pulmonary carcinoma in terms of morbidity and mortality (3). As CRC sufferers typically only present with signs and symptoms in the advanced stages, early detection is challenging. Conventional treatment approaches, which include surgery, chemotherapy, and immunotherapy, are all unsatisfactory (4). Thus, it is critical that more efficient biomarkers be identified to allow for the exploitation of additional potential curative targets for CRC.
As an essential trace element, iron maintains the normal metabolic functions of the body (5). It is involved in many redox reactions by regulating the activity of key enzymes and affects both cell proliferation and apoptosis. Excessive amounts of iron in cells can promote the production of lipid peroxides, which can lead to cell death (6). This cell apoptotic process cannot be inhibited by classical apoptosis inhibitors, is closely related to intracellular iron content, and is referred to as ferroptosis. Ferroptosis is a form of cell death in which large amounts of iron accumulate and lipid peroxidation. Ferroptosis is closely related to the pathophysiological processes of many diseases, including tumors, neurological diseases, and iron metabolism diseases (7). Recent study has found that ferroptosis is closely related to tumor cell proliferation (8). Ferroptosis is a novel kind of cell death that differs from autophagy and apoptosis in that it is iron-dependent and reactive oxygen species (ROS)-dependent. It presents as an iron-dependent accumulation of intracellular lipid ROS, ferroptosis can be triggered by various physiological conditions and pathological stresses and plays a key role in tumorigenesis (9). Ferroptosis can lead to a chronic inflammatory state, which is closely related to the development of tumors. To help neighboring tumor cells survive or evade immunity, tumor cells in the blood and tumor-infiltrating immune cells can produce immunosuppressive mediators such as prostaglandin E2 (PGE2), thereby suppressing anti-tumor immunity and ultimately promoting tumor growth (10). As our understanding of ferroptosis has increased, research into its role in digestive system tumors has continued to intensify (11). A recent study showed that ferroptosis is involved in the development of hepatocellular carcinoma (12). RSL3 is a ferroptosis activator that can trigger ferroptosis in CRC cells by inhibiting the expression of GPX4, increasing the expression of transferrin, and promoting the accumulation of intracellular ROS (13). Propofol is a commonly used clinical intravenous anesthetic, which can inhibit the expression of GPX4 in colorectal tumor cells, while increasing the levels of Fe2+ and ROS, thereby stimulating ferroptosis and inhibiting the growth and proliferation of tumor cells (14). Another study found that ferroptosis is significantly suppressed in gastric cancer, leading to reduced tumor growth and reduced sensitivity to cisplatin and paclitaxel (15). Further, recent studies have shown that ferroptosis is closely related to colon cancer (13).
Based on these findings, many researchers view the molecular prognostic model of ferroptosis-related genes as a prognostic biomarker for cancer patients. Previous research on tumor ferroptosis has primarily focused on scanning relevant databases using bioinformatics analyses to identify any abnormally expressed ferroptosis-related genes that are closely associated with malignancies (16-18). Machine learning is the primary way to implement human intelligence and aims to develop algorithms that can automatically learn from data (19). It can use complex arithmetic to capture large datasets with multidimensional variables to obtain high-dimensional, non-linear relationships between clinical characteristics and make outcome predictions. In this study, a machine-learning analysis was conducted to identify the important genes and molecular interaction networks associated with ferroptosis in CRC.
Methods
Data download
The Gene Expression Omnibus (GEO) datasets for CRC were downloaded from the National Center for Biotechnology Information (NCBI). Detailed information (https://www.ncbi.nlm.nih.gov/) about the 3 downloaded datasets is presented in Table 1. In total, 291 ferroptosis-related genes were downloaded from FerrDb (http://www.zhounan.org/ferrdb/current/) and Gene Cards (https://www.genecards.org). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| ID | CRC cases (n) | Normal cases (n) | Origination | Note |
|---|---|---|---|---|
| COADREAD | 380 | 51 | Cancerous and paraneoplastic tissue | Training set |
| GSE32323 | 17 | 17 | Cancerous and paraneoplastic tissue | Training set (excluding cell line samples) |
| GSE113513 | 14 | 14 | Cancerous and paraneoplastic tissue | Validation set |
CRC, colorectal cancer.
Identification of the ferroptosis-related differentially expressed genes (DEGs)
The COADREAD (Colon and Rectal Cancer) dataset comprised 380 CRC samples and 51 normal samples of cancer tissues and para-cancerous tissues, respectively. The gene symbol expression profile data were downloaded from University of California Santa Cruz Xena (https://xenabrowser.net/datapages/) along with the survival information. The dataset was normalized, and differences between the CRC and normal data were then analyzed using the “limma” package (https://bioconductor.org/packages/release/bioc/html/limma.html). The following threshold was set: an adjusted P value <0.05 and a |log fold change (FC)| >1. The 291 ferroptosis-related genes were intersected with the 262 differential genes to obtain the ferroptosis-related differential expressed genes, of which 11 were ferroptosis-related DEGs. The results were then plotted using the “ggplot2” package (https://cran.r-project.org/web/packages/ggplot2/index.html).
The gene identification expression profile data of the 2 datasets (i.e., GSE32323 and GSE113513) were downloaded from the NCBI and annotated into GeneSymbol using the GPL570 annotation file (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32323; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113513). The datasets were then normalized, and grouped as CRC or normal using the “limma” package for the analysis of variance. The following threshold was set: an adjusted P value <0.05 and a |log FC| >1.
Correlation analysis
The expression profiles of the ferroptosis-related DEGs from the COADREAD dataset were extracted, and the correlations between the genes were calculated using the “corr function” in R (20). The Spearman algorithm (https://rpubs.com/aaronsc32/spearman-rank-correlation) was used. The “ggcorrplot” package was then used to create the plots. The expression profiles of the ferroptosis-related DEGs from the COADREAD dataset were calculated and plotted. The Pearson correlation algorithm (http://www.sthda.com/english/wiki/correlation-test-between-two-variables-in-r) was used. The following correlation threshold was set: a correlated value >0.7 and a P value <0.05.
Machine learning
The ferroptosis-related DEG profiles in the COADREAD dataset were extracted, and the samples were divided into the training and test sets using an 8:2 ratio. The least absolute shrinkage and selection operator (LASSO) regression model was also constructed using the “glmnet” package (https://cran.r-project.org/web/packages/glmnet/index.html) to identify the characteristic genes with non-zero coefficients (16). The ferroptosis-related DEG profiles from the COADREAD dataset were extracted, a 5-fold cross-validation was performed using the R language “rfeControl” package (https://search.r-project.org/CRAN/refmans/caret/html/rfeControl.html), and support vector machine (SVM) models were constructed using the “Caret” package(https://cran.r-project.org/web/packages/caret/index.html) (21). The hub genes obtained from both the SVM and LASSO methods were taken to be the intersection of the total hub genes.
Verification of the hub genes
The validation was performed using the deep-learning method previously described by Tian et al. (22). For the hub genes, their gene expression profiles in the COADREAD dataset and the corresponding sample grouping information were extracted. A multi-layer perceptron (MLP) neural network was constructed using the “neuralnet” package (https://cran.r-project.org/web/packages/neuralnet/index.html) of R language. The constructed neural network models were validated in the 2 datasets of GSE32323 and GSE113513, respectively. Receiver operating characteristic (ROC) curves were used to demonstrate the diagnostic capabilities of the models.
Genetic interaction networks
As described previously by Yu et al. (23), the RNAInter database (http://www.rnainter.org/) was used to screen the pairs of the hub genes that interacted with the micro-RNAs (miRNAs). The RNAInter database was then used to screen the pairs of 2 hub genes that were intercrossed with the long non-coding RNAs (lncRNAs). Interaction pairs with scores >0.5 were considered significant (24), and the pairs were merged and plotted with Cytoscape (https://cytoscape.org/), which is an open-source software platform for visualizing complex networks.
Immunocyte infiltration
The CIBERSORT algorithm (25) was used to identify the immune infiltrates, and the LM22 feature matrix files (25) were used to compare the infiltration of the immune cells in the CRC and normal control samples. The correlations between the hub gene expression of the COADREAD dataset and the immune-related gene set were calculated using the “corr.test” function of Spearman’s algorithm (https://rpubs.com/aaronsc32/spearman-rank-correlation), and the correlation heatmap was drawn. The immune checkpoint genes reported by Chen et al. (26) were used to calculate and plot the correlations between these genes and the hub genes in the COADREAD dataset.
Statistical analysis
For the COADREAD samples that contained survival time and survival status data, a survival curve analysis was conducted using the “survfit” function of the “survival” package (https://cran.r-project.org/web/packages/survival/index.html), and the results were plotted using the “ggsurvplot” function of the “survminer” package (https://cran.r-project.org/web/packages/survminer/readme/README.html). The hub genes were grouped as having high and low expression based on the median for all the samples. Overall survival (OS) and disease-specific survival (DSS) curves were plotted for each of the 4 hub genes. The outcome indicator for OS was the time to death. The outcome indicator for DSS was death due to a specific disease.
Results
Ferroptosis-related DEG identification among the 3 datasets
The COADREAD dataset comprised 992 DEGs, of which 389 were upregulated and 603 were downregulated. In the COADREAD dataset, 291 ferroptosis-related genes were obtained from FerrDb and GeneCards and intersected with the differential results of the overall genes, and 11 ferroptosis-related DEGs were identified (Figure 1).
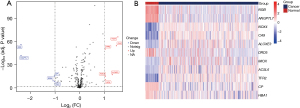
The GSE32323 dataset comprised 1,396 DEGs, of which 682 were upregulated and 714 were downregulated. The GSE32323 data comprised 1,076 DEGs, of which 426 were upregulated and 650 were downregulated. The DEGs associated with iron apoptosis in both the datasets are shown in Table S1.
Correlation analysis of the ferroptosis-related DEGs
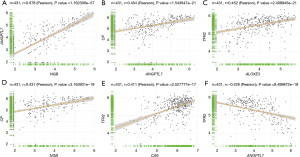
Figure 2 shows some relationships between 2 of the ferroptosis-related genes. The results of the Pearson’s correlation analysis showed that angiopoietin-related protein 7 (ANGPTL7) gene expression was positively correlated with both the neuroglobin (NGB) (r=0.678, Figure 3A) and ceruloplasmin (CP) (r=0.454, Figure 3B) genes. TFR2 gene expression was positively correlated with the arachidonate lipoxygenase 3 (ALOXE3) (r=0.452, Figure 3C). NGB gene expression was positively correlated with the CP (r=0.431, Figure 3D). Transferrin receptor 2 (TFR2) expression was positively correlated with the carbonic anhydrase 9 (CA9) (r=0.411, Figure 3E) but negatively correlated with ANGPTL7 (r=−0.426, Figure 3F).

Machine learning
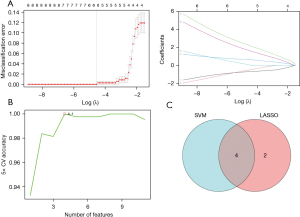
The LASSO regression model identified a total of 6 non-zero coefficient signature genes [i.e., NGB, ANGPTL7, NADPH oxidase 4 (NOX4), CA9, ALOXE3, and TFR2]. Moreover, the SVM model was plotted to compare the accuracy to the number of features, and the number of features corresponding to the point with the highest accuracy was extracted (Figure 4A). As Figure 4B shows, when the optimum number of features was 4, the corresponding 4 optimal signature component genes were NOX4, TFR2, ALOXE3, and CA9 (Figure 4B). As the Venn diagram shows, a total of 4 hub genes were identified; that is, NOX4, TFR2, ALOXE3, and CA9 (Figure 4C).
Verification of the hub genes
A MLP neural network was constructed with the following network structure: 2 hidden layers (Figure 5A). The predicted area under the curve (AUC) value of the GSE32323 dataset was 0.958 (Figure 5B) and the AUC value of the GSE113513 dataset was 0.949 (Figure 5C), which demonstrated excellent diagnostic ability.

Genetic interaction networks
The RNAInter database was used to screen the reciprocal pairs of the hub genes and miRNAs, and a total of 27 miRNAs were identified (22). From the RNAInter database, a total of 193 lncRNAs were screened for interactions with the 2 hub genes (Figure 6).

Immunocyte infiltration
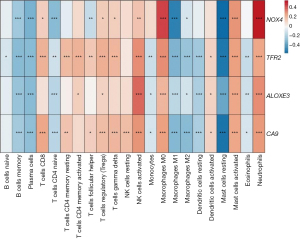
The CIBERSORT algorithm revealed a remarkable difference in the immune cell infiltration in the CRC samples than the normal samples (Figure 7). No significant differences were found in the naive B cells, eosinophils, monocytes, and resting natural-killer cells between the CRC and normal samples.

The expression of the NOX4 gene was elevated and significantly positively correlated with neutrophil (r=0.543) and M0 macrophage (r=0.422) infiltration. In addition, a positive correlation was found between ALOXE3 and activated natural-killer cells (r=0.356, P<0.001). Conversely, NOX4, TFR, and CA9 were negatively correlated with the resting mast cells and had r values of –0.590, –0.536, and –0.546, respectively. For more details on the relationships between the hub genes and immunocyte infiltrations (Figure 8).
The results of the immune checkpoint analysis revealed a strong positive correlation between NOX4 and CD160 expression (r=–0.510, P<0.01) and a significant negative correlation between NOX4 and transforming growth factor beta receptor 1 (TGFBR1) expression (r=0.397, P<0.01, Figure 9).

Statistical analysis
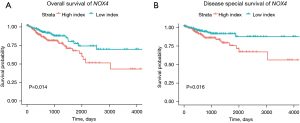
The survival curves for each of the 4 hub genes were plotted separately. Only NOX4 was statistically significant for OS (P=0.014, Figure 10A) and DSS (P=0.016, Figure 10B). These findings suggest that patients had a more favorable prognosis when NOX4 expression levels were relatively low.
Discussion
Presently, operative resection, chemotherapy, targeted-therapies, and immunotherapies dominate the treatment of CRC (27,28). The selection of therapeutic strategies is determined by each patient’s medical status and the clinical characteristics and phase of the tumor. Sadly, not all these alternatives are optimal for advanced CRC patients. To optimize therapeutic clinical regimens, more effective prognostic markers or curative targets need to be identified. Ferroptosis is a new-found model of cell mortality that has been shown to occur in numerous carcinomas and may be of great use in anti-neoplastic therapies (29,30). Ferritin triggers, whether used as mono-therapeutic drugs or in correlation with other chemo-therapeutic drugs, are capable of inhibiting ferritin generation in oncogenic cells, especially in treatment-resistant carcinoma cells (31,32). Ferroptosis may also be selectively aimed at invasive and aggressive cancer stem cells and thus offer the prospect of boosting the effectiveness of immunotherapy and drug tolerance to immunotherapy (33,34). Thus, ferroptosis-related DEGs and the proteins they express represent potentially appropriate anti-cancer interventions for CRC, particularly for patients in the advanced stages or those with treatment resistance.
We identified 11 ferroptosis-related DEGs in the COADREAD dataset. We found that ANGPTL7 gene expression was positively correlated with both the NGB (r=0.678) and CP (r=0.454) genes but was negatively correlated with TFR2 expression (r=–0.426). In addition, TFR2 gene expression was positively correlated with both the ALOXE3 (r=0.452) and CA9 (r=0.411) genes. A total of 4 hub genes were identified by machine learning; that is, NOX4, TFR2, ALOXE3, and CA9. The expression of the NOX4 gene was significantly and positively correlated with neutrophil (r=0.543) and M0 macrophage (r=0.422) infiltration. In addition, a positive correlation was found between ALOXE3 and activated natural-killer cells (r=0.356). The NOX4, TFRs, and CA9 genes were negatively correlated with resting mast cells. The results of the immune checkpoint analysis revealed a strong positive correlation between NOX4 and CD160 expression (r=0.510) and a significant negative correlation between NOX4 and TGFBR1 expression (r=−0.397). The patients had a more favorable prognosis when the NOX4 expression levels were relatively low.
As a member of the nicotinamide adenine dinucleotide phosphate (NADPH) oxygenase family, NOX4 is inducible by ischemia and hypoxia as cells are exposed to ROS (35). It has been suggested that ROS are engaged in many critical cellular processes, such as proliferation, DNA damage response, and angiogenesis, which are clearly relevant to oncogenesis (36). NOX4 is an essential ROS-generating enzyme, and is thus extremely interesting to investigators. NOX4 has been shown to facilitate cell migrations in mammary and pancreatic carcinomas (37-39). NOX4 has also been shown to mediate the cell cycle and facilitate carcinogenesis in melanomas (40). Previous research has also confirmed that NOX4, a ferroptosis-related gene, is a valid biological marker for gastric carcinogenesis (41). NOX4 has been found to be strongly expressed in colon cancer and correlated to an unfavorable outcome (42). Consistent with our results, a previous study showed that NOX4 was significantly enriched in oncogenic-related pathways and clearly correlated with the immune cellular subtypes (43).
We also found a strong negative correlation between NOX4 and CD160 expression, and a positive correlation between TGFBR1 and NOX4 expression. CD160 is a 27-kDa glycoprotein that is strongly correlated with natural-killer cells and CD8+ T lymphocytes with effector cytolytic activities in the peripheral blood (44). The transmission of transforming growth factor beta 1 (TGFB1) signaling from the subcellular surface to the plasma modulates an enormous number of pathological and physiological events (45). The above relationships need to be further explored in the near future. These mentioned relationships can further confirm and enhance the genetic map associated with ferroptosis and may lead to novel medical applications that might be advantageous for immunotherapy and the survival of CRC patients in the long term.
Conclusions
This study identified the ferroptosis-related DEGs of NOX4, TFR2, ALOXE3 and CA9 in CRC and further validated their relationship with immune cell infiltration and associated immune checkpoints. Our findings confirm the influence of the immune microenvironment on CRC. Low NOX4 levels were found to be more favorable to patient outcomes. Our findings may facilitate future clinical diagnoses and outcome assessments of CRC.
Acknowledgments
Funding: None.
Footnote
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-405/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-405/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ahmed M. Colon Cancer: A Clinician's Perspective in 2019. Gastroenterology Res 2020;13:1-10. [Crossref] [PubMed]
- Wang J, Li S, Liu Y, et al. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med 2020;9:361-73. [Crossref] [PubMed]
- Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783-91. [Crossref] [PubMed]
- Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021;325:669-85. [Crossref] [PubMed]
- Fairweather-Tait SJ. Iron. J Nutr 2001;131:1383S-6S. [Crossref] [PubMed]
- Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol 2016;26:165-76. [Crossref] [PubMed]
- Zhang JJ, Du J, Kong N, et al. Mechanisms and pharmacological applications of ferroptosis: a narrative review. Ann Transl Med 2021;9:1503. [Crossref] [PubMed]
- Morales M, Xue X. Targeting iron metabolism in cancer therapy. Theranostics 2021;11:8412-29. [Crossref] [PubMed]
- Su Y, Zhao B, Zhou L, et al. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett 2020;483:127-36. [Crossref] [PubMed]
- Chen M, Zhou X, Chen D, et al. Research progress of ferroptosis in tumors. Chinese Bulletin of Life Sciences 2022;34:1108-15.
- Song Y, Yang H, Lin R, et al. The role of ferroptosis in digestive system cancer. Oncol Lett 2019;18:2159-64. [Crossref] [PubMed]
- Nie J, Lin B, Zhou M, et al. Role of ferroptosis in hepatocellular carcinoma. J Cancer Res Clin Oncol 2018;144:2329-37. [Crossref] [PubMed]
- Sui X, Zhang R, Liu S, et al. RSL3 Drives Ferroptosis Through GPX4 Inactivation and ROS Production in Colorectal Cancer. Front Pharmacol 2018;9:1371. [Crossref] [PubMed]
- Zhao X, Chen F. Propofol induces the ferroptosis of colorectal cancer cells by downregulating STAT3 expression. Oncol Lett 2021;22:767. [Crossref] [PubMed]
- Zhang H, Deng T, Liu R, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer 2020;19:43. [Crossref] [PubMed]
- Hong Z, Tang P, Liu B, et al. Ferroptosis-related Genes for Overall Survival Prediction in Patients with Colorectal Cancer can be Inhibited by Gallic acid. Int J Biol Sci 2021;17:942-56. [Crossref] [PubMed]
- Huang H, Dai Y, Duan Y, et al. Effective prediction of potential ferroptosis critical genes in clinical colorectal cancer. Front Oncol 2022;12:1033044. [Crossref] [PubMed]
- Xu S, Zhou Y, Luo J, et al. Integrated Analysis of a Ferroptosis-Related LncRNA Signature for Evaluating the Prognosis of Patients with Colorectal Cancer. Genes (Basel) 2022;13:1094. [Crossref] [PubMed]
- Huang JX, Shi J, Ding SS, et al. Deep Learning Model Based on Dual-Modal Ultrasound and Molecular Data for Predicting Response to Neoadjuvant Chemotherapy in Breast Cancer. Acad Radiol 2023; Epub ahead of print. [Crossref] [PubMed]
- Duan Y, Yu C, Yan M, et al. m6A Regulator-Mediated RNA Methylation Modification Patterns Regulate the Immune Microenvironment in Osteoarthritis. Front Genet 2022;13:921256. [Crossref] [PubMed]
- Yu R, Zhang J, Zhuo Y, et al. Identification of Diagnostic Signatures and Immune Cell Infiltration Characteristics in Rheumatoid Arthritis by Integrating Bioinformatic Analysis and Machine-Learning Strategies. Front Immunol 2021;12:724934. [Crossref] [PubMed]
- Tian Y, Yang J, Lan M, et al. Construction and analysis of a joint diagnosis model of random forest and artificial neural network for heart failure. Aging (Albany NY) 2020;12:26221-35. [Crossref] [PubMed]
- Yu H, Guo W, Liu Y, et al. Immune Characteristics Analysis and Transcriptional Regulation Prediction Based on Gene Signatures of Chronic Obstructive Pulmonary Disease. Int J Chron Obstruct Pulmon Dis 2021;16:3027-39. [Crossref] [PubMed]
- Lu Q, Ren S, Lu M, et al. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genomics 2013;14:651. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Chen F, Fan Y, Liu X, et al. Pan-Cancer Integrated Analysis of HSF2 Expression, Prognostic Value and Potential Implications for Cancer Immunity. Front Mol Biosci 2021;8:789703. [Crossref] [PubMed]
- Guo S, Zhang E, Zhang B, et al. Identification of Key Non-coding RNAs and Transcription Factors in Calcific Aortic Valve Disease. Front Cardiovasc Med 2022;9:826744. [Crossref] [PubMed]
- Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361-75. [Crossref] [PubMed]
- Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer 2022;22:381-96. [Crossref] [PubMed]
- Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol 2021;18:280-96. [Crossref] [PubMed]
- Kim EH, Shin D, Lee J, et al. CISD2 inhibition overcomes resistance to sulfasalazine-induced ferroptotic cell death in head and neck cancer. Cancer Lett 2018;432:180-90. [Crossref] [PubMed]
- Liu X, Zhang Y, Wu X, et al. Targeting Ferroptosis Pathway to Combat Therapy Resistance and Metastasis of Cancer. Front Pharmacol 2022;13:909821. [Crossref] [PubMed]
- Shibata Y, Yasui H, Higashikawa K, et al. Erastin, a ferroptosis-inducing agent, sensitized cancer cells to X-ray irradiation via glutathione starvation in vitro and in vivo. PLoS One 2019;14:e0225931. [Crossref] [PubMed]
- Wang W, Green M, Choi JE, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270-4. [Crossref] [PubMed]
- Noreng S, Ota N, Sun Y, et al. Structure of the core human NADPH oxidase NOX2. Nat Commun 2022;13:6079. [Crossref] [PubMed]
- Szanto I. NADPH Oxidase 4 (NOX4) in Cancer: Linking Redox Signals to Oncogenic Metabolic Adaptation. Int J Mol Sci 2022;23:2702. [Crossref] [PubMed]
- Tobar N, Guerrero J, Smith PC, et al. NOX4-dependent ROS production by stromal mammary cells modulates epithelial MCF-7 cell migration. Br J Cancer 2010;103:1040-7. [Crossref] [PubMed]
- Boudreau HE, Casterline BW, Rada B, et al. Nox4 involvement in TGF-beta and SMAD3-driven induction of the epithelial-to-mesenchymal transition and migration of breast epithelial cells. Free Radic Biol Med 2012;53:1489-99. [Crossref] [PubMed]
- Messex JK, Adams KLA, Hawkins WG, et al. Oncogenic Kras-Mediated Cytokine CCL15 Regulates Pancreatic Cancer Cell Migration and Invasion through ROS. Cancers (Basel) 2022;14:2153. [Crossref] [PubMed]
- Zhang X, Li H, Liu C, et al. Role of ROS-mediated autophagy in melanoma Mol Med Rep 2022;26:303. (Review). [Crossref] [PubMed]
- Xiao R, Wang S, Guo J, et al. Ferroptosis-related gene NOX4, CHAC1 and HIF1A are valid biomarkers for stomach adenocarcinoma. J Cell Mol Med 2022;26:1183-93. [Crossref] [PubMed]
- Lin XL, Yang L, Fu SW, et al. Overexpression of NOX4 predicts poor prognosis and promotes tumor progression in human colorectal cancer. Oncotarget 2017;8:33586-600. [Crossref] [PubMed]
- Yang X, Yu Y, Wang Z, et al. NOX4 has the potential to be a biomarker associated with colon cancer ferroptosis and immune infiltration based on bioinformatics analysis. Front Oncol 2022;12:968043. [Crossref] [PubMed]
- Oumeslakht L, Aziz AI, Bensussan A, et al. CD160 receptor in CLL: Current state and future avenues. Front Immunol 2022;13:1028013. [Crossref] [PubMed]
- Liu Q, Zhang L, Zhu MS, et al. High-throughput screening on cochlear organoids identifies VEGFR-MEK-TGFB1 signaling promoting hair cell reprogramming. Stem Cell Reports 2021;16:2257-73. [Crossref] [PubMed]


