
Deoxythymidylate kinase (DTYMK) participates in cell cycle arrest to promote pancreatic adenocarcinoma progression regulated by miR-491-5p through TP53 and is associated with tumor immune infiltration
Highlight box
Key findings
• Reduced deoxythymidylate kinase (DTYMK) expression may be considered a novel prognostic biomarker for pancreatic adenocarcinoma (PAAD) patients.
What is known and what is new?
• DTYMK may have a direct impact on the growth and spread of cancer.
• This study primarily focuses on the role of DTYMK expression in predicting the survival rate of patients with PAAD.
What is the implication, and what should change now?
• This research provides additional evidence for its role in PAAD development and point to an attractive approach for research into PAAD-based cancer treatments.
Introduction
Pancreatic adenocarcinoma (PAAD) is a particularly severe form of digestive pancreatic cancer (1) that is characterized by rapid onset, poor prognosis, and high mortality (2). In particular, the pancreas is rich in blood supply and has no capsule, so it is easy for the tumor cells to reach the lymph nodes around the pancreas and even reach tissues and organs such as the liver and lungs in the early stage (3). Pancreatic cancer is difficult to detect in the early stage. The 5-year survival rate is less than 10% (4), and many patients are diagnosed in the middle to late stages, with lymph node metastases or distant metastasis. Recent research has shown that pathogenic pathways are primarily associated with pancreatic cancer onset and progression, such as detachment from the primary lesion, avoidance of immune surveillance, and anchoring growth through lymphatic and blood vessel migration. It is also a multigene-involved and multistep process (5). Currently, the treatment of PAAD patients includes surgery, chemotherapy, radiotherapy, emerging immunotherapy, targeted therapy, etc., but the long-term prognosis is poor. As a result, continued investigation into the pathophysiology and progression of PAAD, as well as the identification of novel treatment targets, is of the highest concern.
In recent years, with the development and application of high-throughput sequencing (NGS), high-resolution mass spectrometry and bioinformatics, a large number of studies on biological diagnosis of pancreatic cancer have emerged, including gene mutation, epigenetic changes, new biomarkers of changes in protein and non-coding RNA levels, such as Luo et al. (6), The sensitivity of CEA to Lewis negative pancreatic cancer patients was higher than that of Lewis positive patients, 63.8% and 42.6%, respectively; The mutation rate of KRAS in pancreatic cancer tissue was 74.7%, and the mutation rate of KRAS in cell-free DNA (cfDNA) was 62.6%. The coincidence rate between the two was 77.3%, indicating that cfDNA can reflect the gene mutation in cancerous tissue to some extent (7). Quantitative methylation specific PCR (qMSP) was used to detect pancreatic cytological samples from patients with pancreatic cancer. It was found that the sensitivity of hypermethylation CDO1 in the diagnosis of pancreatic cancer was 95%, which was more sensitive than endoscopic retrograde cholangiopancreatography aspiration cytology (33%) and EUS-FNA cytology (88%) (8). Zou et al. (9) found six significantly up-regulated miRNAs in the serum of pancreatic cancer patients: let-7b-5p, miR192-5p, miR-19a-3p, miR-19b-3p, miR223-3p and miR-25-3p. And based on these six miRNAs, a serum miRNA detection group was established. However, these biomarkers have more or less deficiencies in specificity, accuracy, or economic costs, and the effectiveness of promotion and implementation is poor. Therefore, further exploration of new biomarkers is needed.
The enzyme deoxythymidylate kinase (DTYMK) plays a critical role in regulating nucleoside thymidine synthesis. It further controls DNA synthesis, replication, and nucleotide metabolism by regulating the synthesis of dTTP (10,11). Furthermore, it is also involved in the first step of the salvage and de novo pathways to produce dTTP (12,13). Recent research has shown a crucial function for DTYMK in embryonic development, specifically in the formation of the nervous system (14). Children with biallelic mutations in DTYMK are prone to severe early-onset neurodegenerative diseases, manifested as microcephaly, growth retardation, brain atrophy (15), impaired DNA replication in fibroblasts and impaired proliferation of brain cells (16). Studies have also shown that DTYMK may have a direct impact on the growth and spread of cancer. For instance, DTYMK expression correlates with immune infiltration in LUAD and modulates cell migration, proliferation, and the cell cycle in lung adenocarcinoma in vitro (17). Inhibition of DTYMK may considerably suppress the development of HCC and enhance sensitivity to oxaliplatin (11). However, there is currently no relevant research on DTYMK in PAAD.
Ultimately, this study primarily focuses on the role of DTYMK expression in predicting the survival rate of patients with PAAD based on data from several datasets, including The Cancer Genome Atlas (TCGA). We further verified our findings using bioinformatics to better understand DTYMK’s impact on tumor cell behavior and its possible mode of action. Our findings will help guide novel approaches to PAAD therapy in clinical practice. We present this article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-393/rc).
Methods
Data analysis using the TCGA database
In the current study, DTYMK expression was evaluated in PAAD cancer and paracancerous tissues, and DTYMK expression was further studied in a pancancer context based on the TCGA database. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DTYMK and PAAD clinical parameters in the TCGA database
Additionally, the clinical parameters of patients with PAAD were downloaded from the TCGA database, and we compared and evaluated them to determine and analyze a possible correlation between DTYMK expression and the outcome of PAAD patients.
Construction and evaluation of nomograms
The Alignment Diagram, also known as the Nomogram, is based on multivariate regression analysis. It integrates multiple predictive indicators and uses scaled line segments to plot them on the same plane at a certain scale, thereby expressing the interrelationships between various variables in the predictive model. By constructing a multi factor regression model, based on the contribution of each influencing factor to the outcome variable (the size of the regression coefficient) in the model, each value level of each influencing factor is assigned a score, and then the total score is obtained by adding the scores. Finally, the predicted value of the individual’s outcome event is calculated by converting the relationship between the total score and the probability of occurrence of the outcome event. Finally, we further evaluated the predictive value of the nomogram using calibration curves.
GeneMANIA/String database analysis of THBS1-interacting genes
We used the String database (https://string-db.org/) and GeneMANIA database (http://www.genemania.org) for the analysis of genetic, protein-DNA, and protein-protein interactions, biochemical and physiological reactions, pathways, and genes. We analyzed the related genes interacting with DTYMK from the perspectives of protein domain, protein expression, and phenotype screening and built a protein-protein interaction network.
TIMER-TCGA correlation between DTYMK and immunological cells
Furthermore, using the TCGA and TIMER databases (https://cistrome.shinyapps.io/timer/), we visualized the correlation between DTYMK expression and different immune cells in PAAD by means of stick figures.
Gene Set Enrichment Analysis (GSEA) enrichment analysis
Moreover, we performed functional analysis using the online Metascape tool and included the identified differentially expressed genes for GSEA.
TargetScan predicted DTYMK upstream miRNAs
The TargetScan database was queried to obtain more information on relevant miRNAs that may bind to DTYMK mRNA at their 3'UTR ends in an effort to deduce the regulatory mechanism of DTYMK via upstream genes.
DTYMK correlation test using the starBase database
We also checked the starBase database for a link between potential miRNAs and DTYMK to further investigate the signaling axis that controls upstream DTYMK.
Statistical analysis
Overall, the visualization and statistical analysis of the current study were performed through R (version 3.6.3), and GSEA was employed to delve into the potential cellular mechanism of DTYMK. However, the statistical significance was determined using the log rank test, while the survival rates were calculated through the Kaplan-Meier technique. The aforementioned online database was used to automatically generate statistical analyses for Yu’s research. Log rank P values less than 0.05 and P values less than 0.05 were classified as significant.
Results
DTYMK expression is upregulated in PAAD
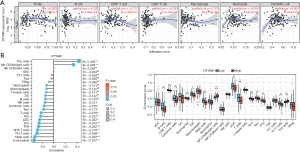
We identified DTYMK in the TCGA database, and our research focused on understanding its role. Through pancancer analysis, we found that DTYMK was differentially expressed in different tumors. There was a substantial (P<0.05) decrease in DTYMK expression in KICH tumor tissues compared to normal tissues (Figure 1A). We searched multiple databases for driver evaluation of DTYMK gene in PanCancer in the TCGA dataset of oncovar.org (https://oncovar.org), and found that some databases suggested that DTYMK gene was a driver gene.However, DTYMK expression was also shown to be greater in unpaired PAAD tumor tissues than in paracancerous tissues (Figure 1B). The receiver operating characteristic (ROC) curve was constructed, and the AUC was found to be 0.677, so DTYMK has a certain predictive research value (Figure 1C).
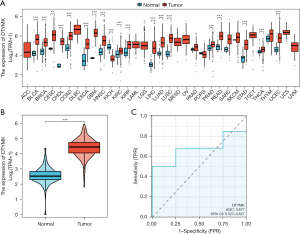
TCGA database DTYMK and PAAD prognostic correlation
We analyzed the clinical data of PAAD from the TCGA database and observed high overall survival (OS), progression free interval (PFI), and disease-specific survival (DSS) in patients with low DTYMK expression compared to patients with high expression (Figure 2A). The results from a subgroup study also indicated that individuals with PAAD who expressed less DTYMK had the best outcomes (Figure 2B).
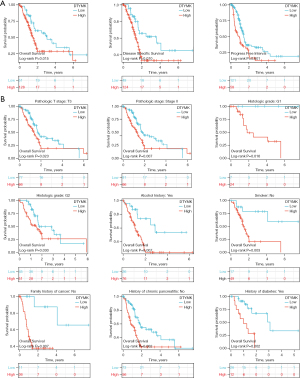
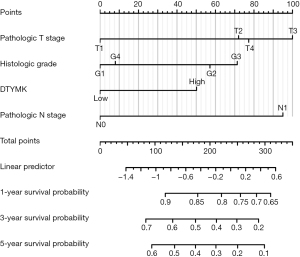
Nomogram construction
On the basis of the multivariate analysis findings, utilizing the TCGA database, we constructed a nomogram to predict the 1-, 3-, and 5-year survival rates of PAAD patients separately; the C-index of the nomogram was 0.644 (0.612–0.677) (Figure 3).
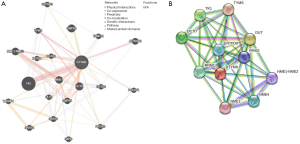
Identification of genes and proteins that interact with DTYMK
Additionally, we mapped out the DTYMK gene network and modified neighboring genes via GeneMania (Figure 4A). Moreover, the DTYMK protein-protein interaction (PPI) network was generated using the STRING database (Figure 4B).
Analysis of DTYMK’s association with immune cells
Consequently, the correlation between DTYMK expression and tumor purity and different immune cells was further demonstrated in PAAD using the TIMER and TCGA databases. Eventually, it was observed that dendritic cells showed a negative association with infiltration, whereas neutrophils, CD4+ T cells, CD8+ T cells, B cells, and macrophages were all found to be favorably correlated with DTYMK expression (Figure 5A). To further understand the influence of DTYMK on the TME, we analyzed its connection with various immune cells and discovered that DTYMK expression was negatively correlated with the infiltration level of most immune cells. This comprised not only Th2 cells and NK CD56bright cells but also mast cells, eosinophils, CD8 T cells, TFH cells, Th17 cells, etc. (Figure 5B). This suggests that DTYMK expression may be linked to PAAD immune infiltration into tumors.
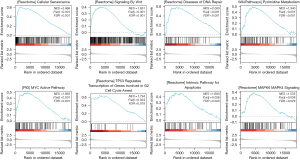
Prediction of signaling pathways based on GSEA
For online functional analysis, we used GSEA and Metascape. Our results suggested that DTYMK’s involvement in cellular senescence, DNA repair, pyrimidine metabolism, MYC activation, TP53 control of cell cycle arrest, apoptosis, and the MAPK6/MAPK4 pathway may influence the biological processes of PAAD and hence influence the prognosis of patients (Figure 6).
DTYMK upstream miRNA prediction from TargetScan
The TargetScan database was used to identify potential miRNAs that bind to DTYMK mRNA at the end of the 3'UTR. Then, the top 10 most likely miRNAs were chosen for further study in an attempt to determine whether DTYMK is controlled by upstream genes. The findings are summarized in Table 1.
Table 1
| miRNA | Position in the UTR | Seed match | Context++ score | Context++ score percentile | Weighted context++ score | Conserved branch length | Pct |
|---|---|---|---|---|---|---|---|
| Hsa-miR-378a-3p | 82–89 | 8mer | −0.42 | 98 | −0.42 | 0 | N/A |
| Hsa-miR-491-5p | 90–96 | 7mer-1A | −0.3 | 95 | −0.3 | 0.083 | N/A |
| Hsa-miR-93-3p | 118–124 | 7mer-1A | −0.2 | 93 | −0.2 | 0.698 | N/A |
| Hsa-miR-181b-3p | 279–285 | 7mer-m8 | −0.21 | 94 | −0.21 | 0 | N/A |
| Hsa-miR-2355-5p | 165–171 | 7mer-m8 | −0.08 | 65 | −0.08 | 0 | N/A |
| Hsa-miR-3199 | 34–40 | 7mer-m8 | −0.21 | 86 | −0.21 | 0 | N/A |
| Hsa-miR-3615 | 41–47 | 7mer-m8 | −0.29 | 87 | −0.29 | 0 | N/A |
| Hsa-miR-545-5p | 269–275 | 7mer-1A | −0.17 | 91 | −0.17 | 0 | N/A |
| Hsa-miR-101-5p | 284–290 | 7mer-m8 | −0.18 | 94 | −0.17 | 0 | N/A |
| Hsa-miR-150-5p | 49–55 | 7mer-1A | −0.06 | 80 | −0.06 | 0.911 | <0.1 |
DTYMK, deoxythymidylate kinase; UTR, Untranslated Regions; Pct, pragmatic clinical trials; N/A, not available.
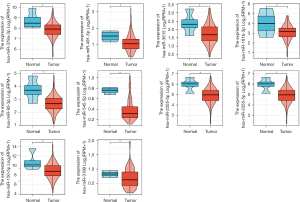
TCGA PAAD candidate miRNA expression analysis
We explored the expression of the above candidate miRNAs in PAAD in the TCGA database and found that hsa-miR-491-5p, hsa-miR-378a-3p, hsa-miR-181b-3p, hsa-miR-93-3p, hsa-miR-101-5p, hsa-miR-3615, hsa-miR-545-5p, hsa-miR-2355-5p and hsa-miR-150-5p were downregulated in PAAD tumor tissues, and the differences were statistically significant (P<0.05) (Figure 7).
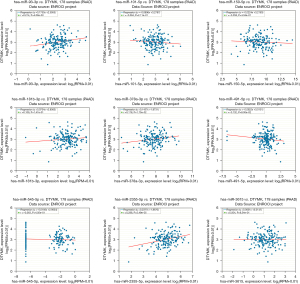
PAAD patient prognosis is correlated with StarBase candidate miRNAs and DTYMK
Subsequently, we further analyzed the association among the above identified miRNAs and DTYMK expression in the StarBase database. The competitive endogenous RNA (ceRNA) hypothesis proposes that long noncoding RNAs (lncRNAs) bind to tumor-suppressing miRNAs, blocking the inhibitory impact of miRNAs on their target mRNAs. Therefore, miRNAs and their target mRNAs should be negatively correlated in the ceRNA network. We found negative correlations between hsa-miR-150-5p, hsa-miR-101-5p, hsa-miR-491-5p and hsa-miR-545-5p and DTYMK expression (Figure 8). Then, we found that patients with high hsa-miR-101-5p, hsa-miR-545, and hsa-miR-491-5p expression had a better prognosis (Figure 9), and this was statistically significant (P<0.05). We conclude that hsa-miR-491-5p is the most likely upstream regulatory miRNA in PAAD based on its expression, correlation strength, and function in the prognosis of PAAD patients (Figure 9).

Discussion
Cancer of the pancreas is one of the most prevalent forms of malignant tumors found in the population. The disease is very malignant and tends to spread to the lymph nodes and liver. Patients typically do not qualify for surgery when they see a doctor, and radiotherapy and chemotherapy are not sensitive enough, which also makes the prognosis poor. For this reason, the pathogenesis of pancreatic cancer is actively being investigated. Understanding the pathogenic mechanism is of great value.
Since the advent of sequencing and omics technologies, there have been more opportunities to investigate potential diagnostic and therapeutic targets and better understand the mechanism behind PAAD (18). Through bioinformatics research, we investigated the function of DTYMK in the development of PAAD as well as potential mechanisms. We discovered that DTYMK expression was upregulated in PAAD patients in the TCGA database and that these patients had improved OS, DSS, PFI, and prognoses when their DTYMK expression was reduced. Additionally, the nomogram created by the predicted outcome of multivariate analysis provides strength to our applied methodology and study molecules for clinical reference. We identified the protein genes interacting with DTYMK through the TCGA database, GeneMania database, and STRING database, which provided a reference for our subsequent mechanistic exploration. In recent years, the emerging immunotherapy has brought hope to improve the survival period of pancreatic cancer. Previous studies have also shown that various immune cells play an important role in the immune microenvironment of pancreatic cancer. For example, activated iNKT cells increase the immunity of NK and T cells and reduce the protective function of Tumor-associated macrophage in the liver metastasis of pancreatic cancer (19). PLA2G2A+cancer related fibroblasts mediate the immune escape of pancreatic cancer by blocking the anti-tumor immune response of CD8+ Cytotoxic T cell (20). Targeting Treg expression of STAT3 enhances NK mediated metastasis monitoring and improves treatment response in pancreatic adenocarcinoma (21). Therefore, we use the method of bioinformatics analysis DTYMK expression was positively correlated with CD4+ T-cell infiltration, CD8+ T-cell infiltration, neutrophil infiltration, B-cell infiltration, and macrophage infiltration but negatively correlated with dendritic cell infiltration and inversely correlated with the infiltration level of most immune cells, as shown by the TIMER database. On the basis of these results, we hypothesize that DTYMK expression is linked to PAAD tumor immune infiltration and that DTYMK plays a crucial role in facilitating tumor cell immune escape in the PAAD tumor microenvironment.
We next used GSEA to determine that DTYMK may influence PAAD-related biological processes by acting on senescence, DNA repair, pyrimidine metabolism, MYC activation, TP53 control of cell cycle arrest, apoptosis, and the MAPK6/MAPK4 pathway. Moreover, certain research investigating PAAD etiology and evolution has verified the disease mechanism. For example, the inhibition of Clusterin inhibits proliferation by inducing cell senescence in pancreatic cancer (22); the capacity of well-differentiated pancreatic endocrine cancers to metastasize is correlated with overexpression of the Met proto-oncogene and insulin-like growth factor binding protein 3 (23); pancreatic cancer cells that overexpress MUC1 are sensitive to glucose restriction and have changes in glutamine metabolism (24); oncogenic KRAS promotes pancreatic cancer progression by regulating nucleotide synthesis (25); pancreatic neuroendocrine tumor cells become resistant to everolimus after MYC overexpression and more sensitive to cyclin-dependent kinase inhibitors (26); dihydrosanguinarine inhibits pancreatic cancer cells by modulating mut-p53/WT-p53 and Ras/Raf/Mek/Erk pathways (27); metformin induces cell cycle arrest and death in QGP-1 pancreatic neuroendocrine tumor cells, hence reducing their ability to proliferate and develop into tumors (28); chloroquine induces apoptosis of pancreatic neuroendocrine tumor cells through endoplasmic reticulum stress (29); and colony formation and cell proliferation in pancreatic cancer are promoted by PMEPA1 through the MAPK signaling pathway (30). Some of these mechanisms are also hot spots in tumor research, such as the direction of Cellular senescence. Cellular senescence is a stable state of terminal cell cycle arrest, which is related to various macromolecule changes, excessive secretion and proinflammatory phenotype. Cell senescence can be regarded as an obstacle to tumorigenesis, so in principle it can constitute the ideal result of any anti-cancer treatment (including pancreatic cancer). Cancer induced aging genes are aging programs driven by activated oncogenic Gene drive. Extracellular mechanisms further enhance the tumor inhibitory function of aging, including metabolic changes and aging related secretory phenotype (SASP). SASP factors, including chemokines, cytokines, growth factors and enzymes, can induce Paracrine signaling senescence or stable proliferation stagnation of adjacent cancer cells. Some SASP factors can also enhance immune monitoring, which in turn can clear aging cells. And our research gene DTYMK also plays an important role in the tumor immune microenvironment of pancreatic cancer. Therefore, this also reflects that our research direction also has good scientific research value and evidence support for theoretical implementation. In addition, we combined the results of the TargetScan, StarBase, and TCGA databases to determine that hsa-miR-491-5p was the most likely possible upstream regulatory miRNA. Currently, hsa-miR-491-5p has also been reported in mechanistic research studies on PAAD and other tumors, such as the apoptosis-inducing microRNA miR-491-5p targeting TP53 and Bcl-XL (31). The miR-491-5p sponge LINC00460 promotes pancreatic cancer progression by sponging miR-491-5p (32). The depletion of tumor suppressor miR-107 in plasma is associated with tumor progression and is a new therapeutic target for pancreatic cancer (33). Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4 (34). Targeting JMJD2B is one way that miR-491-5p has been shown to have an antitumor impact in ERα-breast cancer (35), while targeting this gene by frog oviduct protein hydrolyzate (ORPH) has been shown to increase metastasis in gastric cancer. HCC growth, metastasis, and glycolysis are all slowed by the miR-491-5p/PKM2 axis (36), and the discovery of the FOCAD/miR-491-5p axis has been shown to reduce cancer stemness, drug resistance, and the development of metastases (37). The interaction between linc01615 and miR-491-5p regulates the survival and metastasis of colorectal cancer cells (38). By integrating the above research results, we can boldly speculate that miR-491-5p may negatively regulate DTYMK to participate in cell cycle arrest through TP53 to promote the progression of pancreatic cancer and be associated with tumor immune infiltration, which also provides a basis for our follow-up study. The results of our basic experiments will serve as a guide.
Conclusions
In conclusion, this research demonstrates DTYMK’s potential as a biomarker of PAAD disease progression and provides additional evidence for its role in PAAD development. These results point to an attractive approach for research into PAAD-based cancer treatments.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-393/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-393/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-393/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wan Y, Xu L, Liu Z, et al. Utilising network pharmacology to explore the underlying mechanism of Wumei Pill in treating pancreatic neoplasms. BMC Complement Altern Med 2019;19:158. [Crossref] [PubMed]
- Hosoda W, Wood LD. Molecular Genetics of Pancreatic Neoplasms. Surg Pathol Clin 2016;9:685-703. [Crossref] [PubMed]
- Kato S, Akimoto K, Nagashima Y, et al. aPKCλ/ι is a beneficial prognostic marker for pancreatic neoplasms. Pancreatology 2013;13:360-8. [Crossref] [PubMed]
- Li XZ, Song J, Sun ZX, et al. Diagnostic performance of contrast-enhanced ultrasound for pancreatic neoplasms: A systematic review and meta-analysis. Dig Liver Dis 2018;50:132-8. [Crossref] [PubMed]
- Zhou H, Zhu Y, Wei F, et al. Significance of MUC2 gene methylation detection in pancreatic cancer diagnosis. Pancreatology 2019;19:1049-53. [Crossref] [PubMed]
- Luo G, Liu C, Guo M, et al. Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer. Ann Surg 2017;265:800-5. [Crossref] [PubMed]
- Kinugasa H, Nouso K, Miyahara K, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015;121:2271-80. [Crossref] [PubMed]
- Nishizawa N, Harada H, Kumamoto Y, et al. Diagnostic potential of hypermethylation of the cysteine dioxygenase 1 gene (CDO1) promoter DNA in pancreatic cancer. Cancer Sci 2019;110:2846-55. [Crossref] [PubMed]
- Zou X, Wei J, Huang Z, et al. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med 2019;8:2810-22. [Crossref] [PubMed]
- Wang D, Liu J, Liu S, et al. Identification of Crucial Genes Associated With Immune Cell Infiltration in Hepatocellular Carcinoma by Weighted Gene Co-expression Network Analysis. Front Genet 2020;11:342. [Crossref] [PubMed]
- Sun F, Liu Y, Gong T, et al. Inhibition of DTYMK significantly restrains the growth of HCC and increases sensitivity to oxaliplatin. Cell Death Dis 2021;12:1093. [Crossref] [PubMed]
- Lam CW, Yeung WL, Ling TK, et al. Deoxythymidylate kinase, DTYMK, is a novel gene for mitochondrial DNA depletion syndrome. Clin Chim Acta 2019;496:93-9. [Crossref] [PubMed]
- Arnér ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol Ther 1995;67:155-86. [Crossref] [PubMed]
- Guo Y, Luo W, Huang S, et al. DTYMK Expression Predicts Prognosis and Chemotherapeutic Response and Correlates with Immune Infiltration in Hepatocellular Carcinoma. J Hepatocell Carcinoma 2021;8:871-85. [Crossref] [PubMed]
- He G, Qiu J, Liu C, et al. MiR-148b-3p Regulates the Expression of DTYMK to Drive Hepatocellular Carcinoma Cell Proliferation and Metastasis. Front Oncol 2021;11:625566. [Crossref] [PubMed]
- Yao F, Zhan Y, Li C, et al. Single-Cell RNA Sequencing Reveals the Role of Phosphorylation-Related Genes in Hepatocellular Carcinoma Stem Cells. Front Cell Dev Biol 2021;9:734287. [Crossref] [PubMed]
- Chen X, Yuan Y, Zhou F, et al. Comprehensive analysis of DTYMK for estimating the immune microenvironment, diagnosis, prognosis effect in patients with lung adenocarcinoma. Aging (Albany NY) 2022;14:7866-76. [Crossref] [PubMed]
- Xu D, Wang Y, Zhang Y, et al. Systematic Analysis of an Invasion-Related 3-Gene Signature and Its Validation as a Prognostic Model for Pancreatic Cancer. Front Oncol 2021;11:759586. [Crossref] [PubMed]
- Yi Q, Wang J, Liu T, et al. scRNA-Seq and imaging mass cytometry analyses unveil iNKT cells-mediated anti-tumor immunity in pancreatic cancer liver metastasis. Cancer Lett 2023;561:216149. [Crossref] [PubMed]
- Ge W, Yue M, Lin R, et al. PLA2G2A(+) cancer-associated fibroblasts mediate pancreatic cancer immune escape via impeding antitumor immune response of CD8(+) cytotoxic T cells. Cancer Lett 2023;558:216095. [Crossref] [PubMed]
- Piper M, Van Court B, Mueller A, et al. Targeting Treg-Expressed STAT3 Enhances NK-Mediated Surveillance of Metastasis and Improves Therapeutic Response in Pancreatic Adenocarcinoma. Clin Cancer Res 2022;28:1013-26. [Crossref] [PubMed]
- Mitsufuji S, Iwagami Y, Kobayashi S, et al. Inhibition of Clusterin Represses Proliferation by Inducing Cellular Senescence in Pancreatic Cancer. Ann Surg Oncol 2022;29:4937-46. [Crossref] [PubMed]
- Hansel DE, Rahman A, House M, et al. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res 2004;10:6152-8. [Crossref] [PubMed]
- Gebregiworgis T, Purohit V, Shukla SK, et al. Glucose Limitation Alters Glutamine Metabolism in MUC1-Overexpressing Pancreatic Cancer Cells. J Proteome Res 2017;16:3536-46. [Crossref] [PubMed]
- Santana-Codina N, Roeth AA, Zhang Y, et al. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat Commun 2018;9:4945. [Crossref] [PubMed]
- Terracciano F, Capone A, Montori A, et al. MYC Upregulation Confers Resistance to Everolimus and Establishes Vulnerability to Cyclin-Dependent Kinase Inhibitors in Pancreatic Neuroendocrine Neoplasm Cells. Neuroendocrinology 2021;111:739-51. [Crossref] [PubMed]
- Wu SZ, Xu HC, Wu XL, et al. Dihydrosanguinarine suppresses pancreatic cancer cells via regulation of mut-p53/WT-p53 and the Ras/Raf/Mek/Erk pathway. Phytomedicine 2019;59:152895. [Crossref] [PubMed]
- Yamana H, Kato K, Kobara H, et al. Metformin Inhibits Proliferation and Tumor Growth of QGP-1 Pancreatic Neuroendocrine Tumor Cells by Inducing Cell Cycle Arrest and Apoptosis. Anticancer Res 2020;40:121-32. [Crossref] [PubMed]
- Nakano K, Masui T, Yogo A, et al. Chloroquine induces apoptosis in pancreatic neuroendocrine neoplasms via endoplasmic reticulum stress. Endocr Relat Cancer 2020;27:431-9. [Crossref] [PubMed]
- Song M, Zhou B, Li B, et al. PMEPA1 Stimulates the Proliferation, Colony Formation of Pancreatic Cancer Cells via the MAPK Signaling Pathway. Am J Med Sci 2021;362:291-6. [Crossref] [PubMed]
- Guo R, Wang Y, Shi WY, et al. MicroRNA miR-491-5p targeting both TP53 and Bcl-XL induces cell apoptosis in SW1990 pancreatic cancer cells through mitochondria mediated pathway. Molecules 2012;17:14733-47. [Crossref] [PubMed]
- Wu J, Sun S, Liao W, et al. LINC00460 promotes pancreatic cancer progression by sponging miR-491-5p. J Gene Med 2021;23:e3333. [Crossref] [PubMed]
- Imamura T, Komatsu S, Ichikawa D, et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep 2017;7:5708. [Crossref] [PubMed]
- Yu T, Wang LN, Li W, et al. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci 2018;109:1393-403. [Crossref] [PubMed]
- Hui Z, Yiling C, Wenting Y, et al. miR-491-5p functions as a tumor suppressor by targeting JMJD2B in ERα-positive breast cancer. FEBS Lett 2015;589:812-21. [Crossref] [PubMed]
- Xu Q, Dou C, Liu X, et al. Oviductus ranae protein hydrolysate (ORPH) inhibits the growth, metastasis and glycolysis of HCC by targeting miR-491-5p/PKM2 axis. Biomed Pharmacother 2018;107:1692-704. [Crossref] [PubMed]
- Huang WC, Chi HC, Tung SL, et al. Identification of the Novel Tumor Suppressor Role of FOCAD/miR-491-5p to Inhibit Cancer Stemness, Drug Resistance and Metastasis via Regulating RABIF/MMP Signaling in Triple Negative Breast Cancer. Cells 2021;10:2524. [Crossref] [PubMed]
- Xiao Y, Hu F, Li M, et al. Interaction between linc01615 and miR-491-5p regulates the survival and metastasis of colorectal cancer cells. Transl Cancer Res 2020;9:2638-47. [Crossref] [PubMed]


