
CLEC5A regulates the proliferation and migration of colon cancer via the AKT/mTOR signaling pathway
Highlight box
Key findings
• The expression of CLEC5A was highly elevated in colon cancer tissues and cells and positively associated with malignant pathological progression in colon cancer.
What is known and what is new?
• CLEC5A could mediate the phosphorylation of AKT/mTOR pathway to control colon cancer growth and migration.
• CLEC5A could cooperatively regulate the malignant biological behaviors of colon cancer by interacting with COL1A1 as its target gene.
What is the implication and what should change now?
• Colon cancer patients may benefit from targeting CLEC5A as a novel diagnostic and therapeutic biomarker, which theoretically provides a new direction for preventing drug resistance and tumor recurrence in colon cancer.
Introduction
Colon cancer, the third leading and fourth most deadly malignancy in the world, can be responsible for more than 945,000 new cases and 492,000 death cases annually (1). It is widely known that surgery and chemotherapy are the two main treatment methods for colon cancer, whereas the cancer stages, and tumor location at diagnosis, as well as the individual characteristics of patients are the important reference factors for determining treatment options (2). Even though these standard therapies have been proved to lower cancer morbidity and death, the problems of drug resistance and tumor recurrence are still difficult to overcome (3). Since drug resistance can reportedly develop in nearly all colon cancer patients and limit the therapeutic efficacies of anticancer agents, finally leading to chemotherapy failure (4), colon cancer needs more efficient and accurate screening and treatment options to target potential carcinogenic pathways.
Human C-type lectin domain family 5 member A (CLEC5A), also known as myeloid DAP12-associating lectin-1 (MDL1), has been identified as the II-type transmembrane protein only expressed on bone marrow lineages including monocytes, macrophages, neutrophils, and dendritic cells (DCs) (5-7). Many studies have shown that CLEC5A and other members of this family that share same protein folding pattern are involved in multiple physiological processes, such as cell adhesion, cell growth, turnover, and transfer of glycoproteins (8-10). The main mechanism of this action is that CLEC5A could act as a pattern recognition receptor to transmit signals into cytoplasm through non-covalent binding to the adapter protein DAP12, followed by the phosphorylation of DAP12 initiating a Syk kinase-based signaling cascade, and ultimately leading to the macrophage activation and release of chemokines and pro-inflammatory cytokines (11). Even though CLEC5A has been proved to trigger the bone marrow cell-related immune responses and involve in diverse infections and inflammatory diseases, it is also closely related to the cancer development and progression (12-15).
In actuality, there are studies have suggested that abnormal high CLEC5A expression is significantly correlated with a decrease overall survival time in gastric cancer, glioma, and high-grade severe ovarian cancer (HGSOC) (16-18). At the cellular level, CLEC5A knockdown or silencing could decrease the cell growth and colony formation, as well as the volume and weight of tumor in gastric cancer and brain glioblastoma (16,19). In addition, CLEC5A has also been determined as a potential key gene affecting ovarian cancer’s immune microenvironment remodeling, which could play a role in promoting the immunotherapeutic effect of ovarian cancer patients (20). However, the regulatory mechanism of CLEC5A in colon cancer’s progression is still unknown.
In previous studies, CLEC5A had been proved to promote the tumorigenesis of gastric cancer and brain glioblastoma by activating the PI3K/AKT pathway (16,19), suggesting that CLEC5A may play an important role in malignancies’ incidence and progression. Therefore, we speculate that CLEC5A could also mediate this pathway to regulate the proliferation and migration of colon cancer. Since the extracellular matrix (ECM) acts as a repository for growth factors and a tissue barrier for tumor metastasis, in which the collagen is major structural protein, collagen type I alpha 1 (COL1A1) gene encoding pro-alpha 1 chains of type I collagen could reportedly promote the tumorigenesis and progression of malignancies (21-24). Additionally, COL1A1 has also been identified as a target gene of other genes in promoting the malignant progression of gastric cancer, colon cancer, breast cancer, and lung cancer (25-29), and is closely linked to prognosis of cancer patients (30). Thus, we have reason to speculate that CLEC5A might jointly regulate the proliferation and migration of colon cancer with COL1A1, and notice that there has been no exact study to confirm the interaction between them.
Herein, our research aims to examine CLEC5A expression and function in individuals with colon cancer, especially its correlation with multiple clinicopathological features. Moreover, the molecular mechanism of CLEC5A has also been explored in regulation of colon cancer’s occurrence and development, and the downstream gene “COL1A1” is screened and verified by bioinformatics analysis. These results may pave the way for identifying a novel diagnostic and therapeutic target in colon cancer. We present this article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-304/rc).
Methods
Collection of clinical specimens and cell culture
The tissue samples of 100 colon cancer patients undergoing D2 radical surgery at the Yijishan Hospital Affiliated to Wannan Medical College were obtained and analyzed, in which the adjacent cancerous tissues greater than 5 cm from surgical margin are used as control. Surgical specimens were independently identified as colon adenocarcinoma by two senior pathologists, and none of the patients had received preoperative treatment. The Cell Bank of Chinese Academy of Sciences (Shanghai, China) supplied human colon cancer cell lines (HCT116, SW620, HT29, and SW480); the colon epithelial cell line NCM460 was generously donated by the Comprehensive Cancer Center of Drum Tower Hospital Affiliated to Nanjing Medical University. All colon cancer cells were grown in Roswell Park Memorial Institute (RPMI)-1640 media supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) at 5% CO2 and 37 ℃ conditions. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The clinical study was approved by the Scientific research and New Technology IRB of Wannan Medical College Yijishan Hospital (No. 34) and informed consent was taken from all patients. A protocol was prepared before the study without registration.
Immunohistochemical staining
The clinical specimens were collected to fix with 10% formalin immediately, and then the dehydrated and transparent tissues were embedded in paraffin to make 5 µm-thick continuous slices. The CLEC5A primary antibodies (1:100, Abcam, Cambridge, MA, USA) were used to incubate repaired antigen sections after they had been treated with 3% hydrogen peroxide. This followed by treatment with horseradish peroxidase (HRP)-conjugated secondary antibodies (ZSGB-BIO, Beijing, China). When staining process was completed, the percentage of positive cells and staining intensity in each section were assessed by a light microscope (Olympus, Tokyo, Japan). As previously described, the positive cells percentage levels of 1, 2, 3, and 4 corresponded to 0–25%, 26–50%, 51–75%, and 76–100% tagged cells, respectively, whereas staining intensity scores of 0, 1, and 2 indicated no staining, pale yellow, light brown, and brown (16). Moreover, the immunoreactive score of each slice was obtained by multiplying these two indexes as a standard to make a distinction between the group of CLEC5A low expression [0–3] and high expression [4–12].
Quantitative real-time polymerase chain reaction (qRT-PCR)
Clinical samples, nude mice xenografts, and cell lines underwent total RNA extraction using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and then complementary DNA (cDNA) was synthesized using a reverse transcription (RT) kit (Takara, Dalian, China). The qRT-PCR reaction systems were configured according to a qPCR kit manual (Takara, China), and subsequently amplified by a 7500 real-time PCR equipment (Thermo Fisher Scientific, Waltham, MA, USA). Relative expression of CLEC5A was evaluated with the 2−ΔΔCt technique (GAPDH as internal control).
Western blotting analysis
Tissues and cells were incubated for 30 minutes on ice with radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China), and the lysates obtained were further centrifuged at 12,000 g for 10 minutes to isolate the total proteins. The protein concentration was measured by a bicinchoninic acid (BCA) protein assay kit (Beyotime, China). Next, equivalent amount of protein in each well were separated using sodium dodecyl sulphate polyacrylamide gel electrophoresis (12% SDS-PAGE) as described in manufacturer’s instructions (Beyotime, China), and transferred into the nitrocellulose membranes (PALL, Port Washington, WI, USA). Finally, the membranes were treated with primary antibodies (1:1,000) at 4 ℃ overnight after being blocked with 5% skim milk for 1 hour, and we further incubated membranes with HRP-conjugated secondary antibodies for an additional hour. This was followed by using a chemiluminescence imager (Clinx, Shanghai, China) to exposure protein bands. All primary antibodies, like CyclinD1, CyclinE1, PCNA, E-cadherin, N-cadherin, MMP2, MMP9, p-AKT, p-mTOR, p-P70S6K, p-S6, CLEC5A, and GAPDH were purchased from Abcam (USA).
CLEC5A knockdown cell lines construction
The sequence of short hairpin RNA (shRNA) against CLEC5A was 5'-GGTGGAAATTGGAATTATA-3', which was firstly transfected into HT29 and SW480 cells along with control shRNA using LipofectamineTM 3000 reagent (Thermo Fisher Scientific, USA). Then, the stable knockdown of CLEC5A in HT29 cells were generated by a lentivirus vector (GeneChem, Shanghai, China). The knockdown efficiencies of sh-CLEC5A and control shRNA were validated using WB and qRT-PCR respectively, and repeated these two assays independently for three times.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 kit (Abcam, USA) was used to evaluate the proliferative potential of HT29 and SW480 cells. Briefly, cells were seeded at a density of 5×103 per well in 96-well plates, and then either sh-CLEC5A or its control shRNA was transfected into the cells for 24 hours. Next, cells in each well were incubated with a new RPMI-1640 medium containing 10 µL CCK-8 reagent for 2 hours at culture conditions of 5% CO2 and 37 ℃, and the optical density (OD) value at 450 nm in each indicated time point was measured by a microplate reader (Thermo Fisher Scientific, USA).
Colony formation assay
Transfected cells (both sh-CLEC5A and control shRNA) were plated at a density of 200 per well in 6-well plates. After fixing the colonies formed by proliferative cells with 4% paraformaldehyde (PFA) for 20 minutes, the cells were washed thrice with phosphate-buffered saline (PBS). At last, 0.1% crystal violet solution was used to stain the colonies at room temperature for 30 minutes. It was feasible to determine at least 25 cells within colonies under a microscope (Olympus, Japan).
5-Ethynyl-2'-deoxyuridine assay
We used a 5-Ethynyl-2'-deoxyuridine (EdU) kit (Ribo, Guangzhou, China) to perform the EdU test in accordance with manufacturer’s instructions. To begin, the cells were transfected with sh-CLEC5A and its control shRNA, and then plated in 24-well plates followed by treatment with EdU solution for 2 hours. In step 2, the cells treated above were fixed in 4% PFA and subsequently reacted with Apollo® fluorescent dyes (Apollo Scientific, Greater Manchester, UK); whereas the nucleus were stained with Hoechst 33342 reaction solution. Eventually, inverted fluorescent microscope (Olympus, Japan) was used to determine the proportion of EdU positive staining cells (red fluorescence) to Hoechst staining cells (blue fluorescence).
Wound healing assay
To investigate the migration abilities of HT29 and SW480 cells, a wound healing assay was carried out according to the methods previously described (31). Similarly, the cells transfected with sh-CLEC5A and its control shRNA were firstly seeded in 6-well plates and cultured to 100% density. Secondly, we used sterile pipette tips to vertically scratch the monolayer fused cells followed by washing with PBS to discard floating cells. At last, the treated cells were re cultured in new RPMI-1640 medium without FBS for 48 hours. An inverted light microscope was used to determine the scratch healing rate of each indicated time point (0, 24, and 48 hours).
Transwell assay
Polyester membrane cell culture inserts (Corning, NY, USA) were used to perform the transwell assay. Specifically, the bottom chamber of each well was filled with 600 µL RPMI-1640 media supplemented with 20% FBS, and the upper chamber was stocked with 200 µL suspension (RPMI-1640 medium without FBS) containing around 2×105 cells. After 20 minutes of fixation in 4% PFA and 30 minutes of staining in 0.1% crystal violet solution, cells that had migrated to lower layers were gently removed from the chamber surface using cotton swabs. In the end, a light microscope (Olympus, Japan) was used to take images of 3 random fields from each group.
Tumor xenograft mice models
The 4-week-old male BALB/c athymic mice were randomly divided into two groups (n=5), which were used to establish subcutaneous tumor xenograft models. Above all, the injection of approximately 2×106 HT29 cells stably expressing sh-CLEC5A and its control shRNA was made into the left armpit of mouse in each group, respectively. The lengths and widths of tumor xenografts were measured every 3 days to estimate their volumes (length × width2 × 0.5). Eventually, all mice were euthanized by the intravenous injection of sodium pentobarbital in the 18th day, the tumor grafts were then removed, weighed, and stored at –80℃ refrigerator. SPF mice were purchased from Jiangsu Jicui Pharmaceutical Kang Biotechnology Co., Ltd (China). The animal experiments were performed under a project license (No. LLSC-2022-011) granted by Laboratory Animal Welfare and Ethics Committee, Wannan Medical College, in compliance with relevant institutional guidelines for the care and use of animals.
The Cancer Genome Atlas (TCGA) bioinformatics analysis
We firstly verified the levels of CLEC5A expression in multiple human cancers through Oncomine database (https://www.oncomine.org/). After that, the gene expression data of colon cancer was retrieved and downloaded from TCGA database (https://portal.gdc.cancer.gov/), and further screened by the criteria of P<0.05 and log2(FC) >2 to identify up-regulated genes. On the basis of above-mentioned gene expression data, we plotted CLEC5A expression box diagram and conducted a gene set enrichment analysis (GSEA) to examine potential interconnected signaling pathways in colon cancer. The last step was to statistically analyze and visualize the expression association between CLEC5A and COL1A1 in colon cancer using GraphPad Prism 9.0 software (GraphPad Software Inc., San Diego, CA, USA).
Statistical analysis
SPSS 26.0 software (IBM Corp., Armonk, NY, USA) was used for statistical analysis. At least 3 independent experiments were conducted, with quantitative data presented as mean ± standard deviation (SD) and qualitative data as rate and percentage. Differences between two groups were analyzed using Student’s t-test, whereas one-way analysis of variance (ANOVA) was used to compare multiple groups. Additionally, Spearman’s rank correlation test was utilized to examine statistical significance among two groups. Statistical significance was considered at P<0.05.
Results
CLEC5A differential expression in colon cancer tissues and cells
Firstly, Oncomine database was used to conduct a thorough investigation of CLEC5A expression in multiple malignant and precancerous tissues. This identified 6 different kinds of malignancies to have increased CLEC5A expression [carcinoma of the central nervous system (CNS), breast, intestine, stomach, kidney, pancreas, and other tissues] (Figure 1A). Subsequently, TCGA gene expression data and qRT-PCR confirmed that CLEC5A was markedly elevated in colon cancer tissues, which was consistent with the previously observed messenger RNA (mRNA) expression in colon cancer and adjacent normal tissues (Figure 1B,1C). Moreover, the CLEC5A expression levels in 100 pairs of cancerous and para-cancerous tissues collected from the patients diagnosed as colon adenocarcinoma were detected with immunohistochemistry (IHC). Alongside positive links with the lymph node metastasis, vascular metastasis, and tumor-node-metastasis (TNM) stages of colon cancer patients (P<0.05), IHC data also revealed that CLEC5A was overexpressed in colon cancer tissues compared to adjacent normal tissues (Table 1, and Figure 1D). However, we had previously analyzed corresponding clinical follow-up data of colon cancer patients downloaded from TCGA database, suggesting that no significant correlation existed between CLEC5A expression levels and patient’s survival time (Figure 1E), so the survival analysis based on IHC data had not been developed in this study to further explore CLEC5A prognostic value. Eventually, qRT-PCR was used to deeply examine CLEC5A mRNA expression in 4 different kinds of colon cancer cell lines (HCT116, SW620, HT29, and SW480), revealing elevated CLEC5A expression levels in all the 4 types of colon cancer cells relative to colon epithelial cell (NCM460) (Figure 1F). We focused on HT29 and SW480 cells owing to their notably higher levels of CLEC5A mRNA expression than others. Overall, these above results indicated that CLEC5A could play an important role in the carcinogenesis of colon cancer due to its up-regulated expression levels in colon cancer tissues and cells.
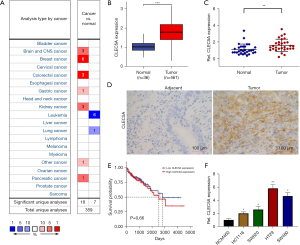
Table 1
| Clinicopathological features | CLEC5A negative (n=41) | CLEC5A positive (n=59) | P value |
|---|---|---|---|
| Age (years) | 0.881 | ||
| <60 | 14 | 21 | |
| ≥60 | 27 | 38 | |
| Gender | 0.243 | ||
| Male | 25 | 29 | |
| Female | 16 | 30 | |
| Pathology type | 0.234 | ||
| Ulcerative type | 31 | 38 | |
| Protruded type | 10 | 21 | |
| Differentiation | 0.308 | ||
| Poor differentiation | 4 | 10 | |
| Moderate differentiation | 37 | 49 | |
| Depth of tumor invasion | 0.132 | ||
| T1–2 | 13 | 11 | |
| T3–4 | 28 | 48 | |
| Lymph node metastasis | 0.026* | ||
| N0 | 33 | 35 | |
| N1–2 | 8 | 24 | |
| Vascular metastasis | 0.031* | ||
| No | 35 | 39 | |
| Yes | 6 | 20 | |
| TNM stages | 0.039* | ||
| I–II stages | 28 | 28 | |
| III–IV stages | 13 | 31 |
*, P<0.05. TNM, tumor-node-metastasis.
CLEC5A knockdown inhibited colon cancer proliferation in vitro
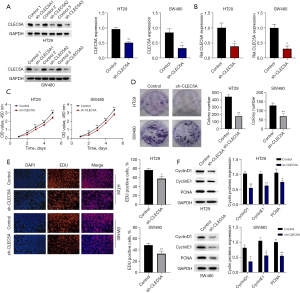
To investigate the effect of CLEC5A on colon cancer proliferation, sh-CLEC5A and its control shRNA were firstly transfected into HT29 and SW480 cells. After constructing the CLEC5A knockdown colon cancer cell models, qRT-PCR and WB assays were used to identify its knockdown efficiency (Figure 2A,2B). Additionally, to further examine these changes in proliferation capacities of HT29 and SW480 cells, we performed the CCK-8, colony formation, and EdU tests. Their results clearly indicated that OD values, colony counts, and EdU positive staining cell proportions were significantly reduced in sh-CLEC5A transfected cells compared to its control (P<0.05) (Figure 2C-2E). Since PCNA had been shown to work in DNA replication and repair, whereas CyclinD1 and CyclinE1 had also been reportedly involve in cell cycle process (32,33), we exploited WB assay to detect the relative expression levels of these proteins in sh-CLEC5A and its control shRNA transfected cells respectively. The results unexceptionally indicated that CLEC5A knockdown substantially suppressed the expression of cell proliferation-related proteins in HT29 and SW480 cells (P<0.05) (Figure 2F). On basis of findings, we hypothesized that CLEC5A could facilitate colon cancer proliferation.
CLEC5A knockdown inhibited colon cancer migration in vitro
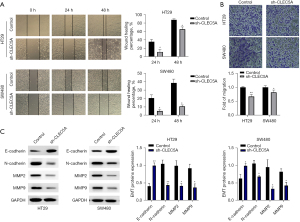
Furthermore, wound healing and transwell tests were conducted in HT29 and SW480 cells transfected with sh-CLEC5A and its control shRNA to deeply examine the role of CLEC5A in colon cancer migration. The results also undoubtedly revealed that wound healing percentages and migrating cell counts were significantly lower in sh-CLEC5A transfected cells than its control (P<0.05) (Figure 3A,3B). Among the complicated and precise molecular mechanisms of colon cancer migration, numerous studies had proved that epithelial-mesenchymal transition (EMT) was one of the most essential pathways in malignant progression (34,35). So once again, we employed WB assay to compare sh-CLEC5A transfected cells to its control for EMT (E-cadherin, N-cadherin, MMP2, and MMP9) proteins expression. Consistently with the previous tests on cell migration capacities, CLEC5A knockdown could dramatically decrease N-cadherin, MMP2, and MMP9 expression, whereas increase E-cadherin expression (P<0.05) (Figure 3C). Thus, these results helped to identify CLEC5A’s promotion in colon cancer migration.
CLEC5A knockdown inhibited colon cancer growth in vivo
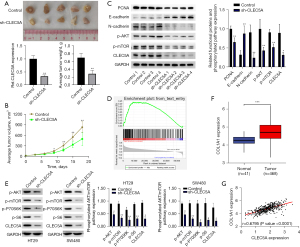
According to the CLEC5A’s extracorporeal regulating role in colon cancer proliferation and migration, we speculated that CLEC5A may also promote tumor growth in vivo, so the nude mice xenografts models were constructed to reveal its effect on colon cancer tumorigenesis. Specifically, the sh-CLEC5A stably transfected HT29 cells were firstly injected into left armpit of each mouse in experimental group, whereas its control cells were equally injected into same site of each control group mouse. Then, the volumes of subcutaneous xenografts were measured and recorded every 3 days to compare tumor growth rates between two groups, followed by euthanasia of all mice on the 18th day to remove xenograft tumors for analysis. Unsurprisingly, the results demonstrated that scales, weights, and growth rates of xenograft tumors were decreased in experimental mice compared to its control (P<0.05) (Figure 4A,4B), and CLEC5A mRNA was also confirmed to have been efficiently knocked down in nude mice xenografts models by qRT-PCR (Figure 4A). Additionally, we employed WB assay to further detect relative levels of several typical cell cycle and EMT-related proteins (PCNA, E-cadherin, and N-cadherin) in randomly selected 4 pairs of tumor grafts from experimental and control mice. The results similarly showed that CLEC5A knockdown could internally reduce PCNA and N-cadherin expression, whereas elevate E-cadherin expression (P<0.05) (Figure 4C). Together, these findings revealed that CLEC5A may promote colon cancer malignant progression by activating the cell cycle and EMT processes in vivo.
CLEC5A knockdown suppressed the AKT/mTOR pathway phosphorylation of colon cancer in vitro and in vivo
Numerous studies had previously determined an indispensable role of the AKT/mTOR pathway phosphorylation in tumorigenesis and progression (36-38), so we speculated that CLEC5A may promote the proliferation and migration of colon cancer by activating this signaling pathway. On basis of TCGA gene expression data, we performed GSEA analysis to validate a positive correlation between CLEC5A expression and AKT/mTOR pathway phosphorylation [normalized enrichment score (NES) =1.37, false discovery rate (FDR) =0.000, P=0.000] (Figure 4D). Subsequently, a WB assay was conducted to ulteriorly examine phosphorylation levels of several AKT/mTOR pathway key proteins (p-AKT, p-mTOR, p-P70S6K, and p-S6) in CLEC5A knockdown colon cancer tissues and cells. Comparing with controls, the CLEC5A knockdown tissues and cells could reduce these phosphorylated proteins expression (P<0.05) (Figure 4C,4E). Therefore, we had reason to believe that CLEC5A may phosphorylate the AKT/mTOR pathway to promote colon cancer’s growth and migration.
CLEC5A and COL1A1 interactions co-regulated colon cancer progression
To elaborate the molecular mechanism by which CLEC5A controlled the cell cycle and EMT processes of colon cancer, we conducted bioinformatics analysis in accordance with TCGA gene expression data. Given that COL1A1 had purportedly been linked to the onset and progression of various malignancies (22-30), we speculated that COL1A1 may co regulate colon cancer progression as a target gene of CLEC5A. Consistent with our hypothesis, the colon cancer tissues from TCGA database had considerably higher levels of COL1A1 expression than adjacent normal tissues (P<0.05) (Figure 4F). After that, we also performed correlation analysis to verify the relationship between CLEC5A and COL1A1, whose result undoubtedly revealed the positive link between two genes’ expression (P<0.05) (Figure 4G). In short, these results partially indicated that CLEC5A may interact with COL1A1 to regulate colon cancer’s malignant progression.
Discussion
Colon cancer, as the fourth most deadly cancer worldwide, can reportedly cause nearly 700,000 deaths per year, which has been ranked after lung, liver, and stomach cancer (39). Recent studies have shown that only 50% of colon cancer patients with phase III can be cured by surgery alone, and 20% can be relieved with adjuvant chemotherapy, whereas the remaining 30% may recur within 2–3 years (40). Consequently, we need to have a thorough investigation of colon cancer’s pathogenesis, which could help us to customize reasonable adjuvant treatment schemes for colon cancer.
More recently, numerous studies have defined the vital role of tumor microenvironment (TME) in predicting survival prognosis and evaluating therapeutic efficacy, its value in colon cancer certainly cannot be ignored as well (41). It is widely known that the TME contains not only cancer cells, but also non-cancer components consist of extracellular matrix, blood vessels, immune cells, neurons, and cells with other tissue functions (42). Unexceptionally, these above cells contribute to the formation of malignant phenotypes in cancer, including out-of-control proliferation, resistance to apoptosis, and evasion of immunosurveillance (43). Owing to reports that colon cancer cells could regulate the polarization of tumor-related immune cells, and tumor-related macrophages influenced angiogenesis and metastasis (44), thereby the adaptive immune reaction in colon cancer may be strongly linked to prognosis and recurrence.
Emerging evidences have demonstrated that CLEC5A partake in immune-inflammatory reactions, such as the proinflammation cytokines releasing, macrophage activation, and neutrophil extracellular trap (45-47). As a type II transmembrane protein, CLEC5A has been reportedly not only involved in multiple inflammatory diseases (Crohn’s disease, autoimmune arthritis, and chronic obstructive pulmonary disease), but also associated with cancer immunity and prognosis in a pan-cancer investigation (48-51). Additionally, CLEC5A has been identified as a carcinogenic factor in tumorigenesis and progression, and a potential biomarker for several malignancies’ poor prognoses, like gastric cancer, glioma, and HGSOC (16-18). Therefore, we hypothesize that CLEC5A may also serve as a regulatory role in colon cancer’s occurrence and advancement.
Here, the differential expression of CLEC5A in human malignancies was firstly verified through Oncomine database, particularly the levels in CNS, breast, colorectal, gastric, kidney, and pancreatic cancers, which exactly coincided with the conclusions in earlier studies. To further investigate the role and mechanism of CLEC5A in colon cancer, we subsequently identified its aberrant high expression in colon cancer tissues using TCGA database analysis and qRT-PCR. Moreover, the IHC results also suggested a favorable correlation between CLEC5A expression levels and malignant pathological features of colon cancer patients (the amounts of lymph node metastases, vascular metastasis, and TNM stages), which hinted that CLEC5A may involve in tumorigenesis and progression. Nevertheless, the corresponding follow-up data of colon cancer patients downloaded from TCGA database indicated that CLEC5A’s prognostic impact on the survival time of colon cancer patients was insignificant, which differed with the analogical survival analysis on gastric cancer, glioma, and HGSOC (16-18). Thereby, the prognostic value of CLEC5A based on IHC data had not been explored in our study.
It is no doubt that CLEC5A could involve in the malignant progression of colon cancer, yet its underlying mechanism remains unclear. We consequently implemented the cell function tests in HT29 and SW480 cells to identify CLEC5A’s regulatory role in colon cancer proliferation and migration. Unsurprisingly, the results indicated that CLEC5A knockdown could markedly reduce OD values, colony counts, EdU positive staining cell proportions, wound healing percentages, and migrating cell counts in colon cancer cells, especially inhibited the cell cycle and EMT processes. Moreover, we constructed nude mice xenografts models to support this moderation effect of CLEC5A in vivo, and the result equally confirmed that knocking down CLEC5A could dramatically slow the tumor growth and malignant biological behaviors of colon cancer.
Mechanistically, there is conclusive evidence that CLEC5A facilitates the tumorigenesis and pathogenesis in glioblastoma and gastric cancer through PI3K/AKT/mTOR pathway (16,19). Since numerous tumor characteristics, including cell proliferation, apoptosis, metabolism, and angiogenesis have been linked to PI3K/AKT/mTOR pathway (52,53), we postulate that CLEC5A may also regulate the proliferation and migration of colon cancer by activating this pathway. Hence, the significance of CLEC5A expression and AKT/mTOR pathway phosphorylation was then verified using GSEA and WB analyses, which unambiguously demonstrated that CLEC5A overexpression was associated with elevated phosphorylation in the AKT/mTOR signaling pathway.
Previous studies have proved that CLEC5A can be activated by the pathogen-associated molecular patterns (PAMPs) expressed by microorganisms, and serve as a key pattern recognition receptor (PRR) (10,54). However, the exogenous and endogenous ligands of most members in C-type lectin families, including CLEC5A, have not been identified yet, we predict that CLEC5A may regulate colon cancer’s pathogenesis and progression by interacting with other target genes. Cell adhesion, cell migration, vascular formation, and ECM remodeling are just some of the many complicated biological processes that relevant to collagen type I alpha 1 (COL1A1), the primary component of the ECM (55-57). Furthermore, several malignant tumor types, including lung cancer, ovarian cancer, and nasopharyngeal carcinoma, have been linked to COL1A1’s role as the mediator of PI3K/AKT/mTOR pathway in their tumor evolution (24,58,59). Similar to these findings, the correlation analysis on basis of TCGA data exactly identified the notable positive dependence between CLEC5A and COL1A1 expression levels, whereas the interaction mechanism of these two still needing to be further investigated.
Taking these observations together, our study firstly determined the positive correlation between high levels of CLEC5A expression and malignant pathological characteristics of colon cancer patients at the clinical aspect, and secondly confirmed the suppressive effect of CLEC5A knockdown by cell function tests and nude mice xenografts models at the cellular and zoological aspect, we accordingly supposed that CLEC5A may serve as a positive regulator in colon cancer’s malignant growth and progression. Specifically, we further performed GSEA and WB analyses to investigate the molecular mechanism of CLEC5A, which convincingly proved that CLEC5A could promote the cell cycle and EMT processes by phosphorylating AKT/mTOR pathway in colon cancer. Ultimately, the correlation analysis based on TCGA data also indicated that CLEC5A may play this regulatory role by interacting with COL1A1. Nevertheless, the limitation of our research lies in lacking accurate understanding of CLEC5A overexpression role in colon cancer’s pathogenesis and progression, and the relationship between CLEC5A and COL1A1 was only identified from data analysis, so we need to provide more experimental evidence to construct the regulatory axis of these two in tumor evolution.
Conclusions
In summary, our research not only enhance the theoretical understanding of CLEC5A’s regulatory role in the proliferation and migration of colon cancer, but also illuminate a way towards the prospective targeted therapy for colon cancer patients.
Acknowledgments
Funding: This work was supported by the Key Fund Project of Wannan Medical College in 2020 (No. WK2020ZF17), Medical Research Development Fund Project of Beijing Kangmeng Charity Foundation (No. YXKY-TB206002), Natural Science Research Project of Anhui Provincial Colleges and Universities (No. KJ2020ZD54), and Top academic talents in discipline (specialty) Funded project (No. gxbjzd2021060).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-304/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-304/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-304/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-304/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The clinical study was approved by Scientific research and New technology IRB of Wannan Medical College Yijishan Hospital (No. 34) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. LLSC-2022-011) granted by Laboratory Animal Welfare and Ethics Committee, Wannan Medical College, in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Weitz J, Koch M, Debus J, et al. Colorectal cancer. Lancet 2005;365:153-65. [Crossref] [PubMed]
- Stein A, Atanackovic D, Bokemeyer C. Current standards and new trends in the primary treatment of colorectal cancer. Eur J Cancer 2011;47:S312-4. [Crossref] [PubMed]
- Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature 2019;575:299-309. [Crossref] [PubMed]
- Dallas NA, Xia L, Fan F, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res 2009;69:1951-7. [Crossref] [PubMed]
- Sung PS, Hsieh SL. CLEC2 and CLEC5A: Pathogenic Host Factors in Acute Viral Infections. Front Immunol 2019;10:2867. [Crossref] [PubMed]
- Chen ST, Lin YL, Huang MT, et al. Targeting C-type lectin for the treatment of flavivirus infections. Adv Exp Med Biol 2011;705:769-76. [Crossref] [PubMed]
- Bakker AB, Baker E, Sutherland GR, et al. Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc Natl Acad Sci U S A 1999;96:9792-6. [Crossref] [PubMed]
- Schäffler H, Rohde M, Rohde S, et al. NOD2- and disease-specific gene expression profiles of peripheral blood mononuclear cells from Crohn's disease patients. World J Gastroenterol 2018;24:1196-205. [Crossref] [PubMed]
- Lo YL, Liou GG, Lyu JH, et al. Dengue Virus Infection Is through a Cooperative Interaction between a Mannose Receptor and CLEC5A on Macrophage as a Multivalent Hetero-Complex. PLoS One 2016;11:e0166474. [Crossref] [PubMed]
- Teng O, Chen ST, Hsu TL, et al. CLEC5A-Mediated Enhancement of the Inflammatory Response in Myeloid Cells Contributes to Influenza Virus Pathogenicity In Vivo. J Virol 2017;91:e01813-16. [Crossref] [PubMed]
- Sung PS, Chang WC, Hsieh SL. CLEC5A: A Promiscuous Pattern Recognition Receptor to Microbes and Beyond. Adv Exp Med Biol 2020;1204:57-73. [Crossref] [PubMed]
- Chen ST, Li FJ, Hsu TY, et al. CLEC5A is a critical receptor in innate immunity against Listeria infection. Nat Commun 2017;8:299. [Crossref] [PubMed]
- Humphrey MB, Nakamura MC. A Comprehensive Review of Immunoreceptor Regulation of Osteoclasts. Clin Rev Allergy Immunol 2016;51:48-58. [Crossref] [PubMed]
- Bao Y, Wang L, Shi L, et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett 2019;24:38. [Crossref] [PubMed]
- Marín-Aguilera M, Reig Ò, Lozano JJ, et al. Molecular profiling of peripheral blood is associated with circulating tumor cells content and poor survival in metastatic castration-resistant prostate cancer. Oncotarget 2015;6:10604-16. [Crossref] [PubMed]
- Wang Q, Shi M, Sun S, et al. CLEC5A promotes the proliferation of gastric cancer cells by activating the PI3K/AKT/mTOR pathway. Biochem Biophys Res Commun 2020;524:656-62. [Crossref] [PubMed]
- Tong L, Li J, Choi J, et al. CLEC5A expressed on myeloid cells as a M2 biomarker relates to immunosuppression and decreased survival in patients with glioma. Cancer Gene Ther 2020;27:669-79. [Crossref] [PubMed]
- Wang R, Du X, Zhi Y. Screening of Critical Genes Involved in Metastasis and Prognosis of High-Grade Serous Ovarian Cancer by Gene Expression Profile Data. J Comput Biol 2020;27:1104-14. [Crossref] [PubMed]
- Fan HW, Ni Q, Fan YN, et al. C-type lectin domain family 5, member A (CLEC5A, MDL-1) promotes brain glioblastoma tumorigenesis by regulating PI3K/Akt signalling. Cell Prolif 2019;52:e12584. [Crossref] [PubMed]
- Shen J, Liu T, Lv J, et al. Identification of an Immune-Related Prognostic Gene CLEC5A Based on Immune Microenvironment and Risk Modeling of Ovarian Cancer. Front Cell Dev Biol 2021;9:746932. [Crossref] [PubMed]
- Prockop DJ. Mutations that alter the primary structure of type I collagen. The perils of a system for generating large structures by the principle of nucleated growth. J Biol Chem 1990;265:15349-52. [Crossref] [PubMed]
- Liu J, Shen JX, Wu HT, et al. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med 2018;25:211-23. [PubMed]
- Geng Q, Shen Z, Li L, et al. COL1A1 is a prognostic biomarker and correlated with immune infiltrates in lung cancer. PeerJ 2021;9:e11145. [Crossref] [PubMed]
- Li M, Wang J, Wang C, et al. Microenvironment remodeled by tumor and stromal cells elevates fibroblast-derived COL1A1 and facilitates ovarian cancer metastasis. Exp Cell Res 2020;394:112153. [Crossref] [PubMed]
- Li Y, Sun R, Zhao X, et al. RUNX2 promotes malignant progression in gastric cancer by regulating COL1A1. Cancer Biomark 2021;31:227-38. [Crossref] [PubMed]
- Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol 2018;96:19-25. [Crossref] [PubMed]
- Zhu X, Luo X, Jiang S, et al. Bone Morphogenetic Protein 1 Targeting COL1A1 and COL1A2 to Regulate the Epithelial-Mesenchymal Transition Process of Colon Cancer SW620 Cells. J Nanosci Nanotechnol 2020;20:1366-74. [Crossref] [PubMed]
- Kim K, Kim YJ. RhoBTB3 Regulates Proliferation and Invasion of Breast Cancer Cells via Col1a1. Mol Cells 2022;45:631-9. [Crossref] [PubMed]
- Zhang H, Li X, Jia M, et al. Roles of H19/miR-29a-3p/COL1A1 axis in COE-induced lung cancer. Environ Pollut 2022;313:120194. [Crossref] [PubMed]
- Li X, Sun X, Kan C, et al. COL1A1: A novel oncogenic gene and therapeutic target in malignancies. Pathol Res Pract 2022;236:154013. [Crossref] [PubMed]
- Martinotti S, Ranzato E. Scratch Wound Healing Assay. Methods Mol Biol 2020;2109:225-9. [Crossref] [PubMed]
- González-Magaña A, Blanco FJ. Human PCNA Structure, Function and Interactions. Biomolecules 2020;10:570. [Crossref] [PubMed]
- Li Y, Wang Y, Li J, et al. Tacrolimus inhibits oral carcinogenesis through cell cycle control. Biomed Pharmacother 2021;139:111545. [Crossref] [PubMed]
- Weigel MT, Krämer J, Schem C, et al. Differential expression of MMP-2, MMP-9 and PCNA in endometriosis and endometrial carcinoma. Eur J Obstet Gynecol Reprod Biol 2012;160:74-8. [Crossref] [PubMed]
- Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol 2019;29:212-26. [Crossref] [PubMed]
- Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Semin Cancer Biol 2019;59:147-60. [Crossref] [PubMed]
- Fattahi S, Amjadi-Moheb F, Tabaripour R, et al. PI3K/AKT/mTOR signaling in gastric cancer: Epigenetics and beyond. Life Sci 2020;262:118513. [Crossref] [PubMed]
- Miricescu D, Totan A, Stanescu-Spinu II, et al. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int J Mol Sci 2020;22:173. [Crossref] [PubMed]
- Brody H. Colorectal cancer. Nature 2015;521:S1. [Crossref] [PubMed]
- Auclin E, Zaanan A, Vernerey D, et al. Subgroups and prognostication in stage III colon cancer: future perspectives for adjuvant therapy. Ann Oncol 2017;28:958-68. [Crossref] [PubMed]
- Chen W, Huang J, Xiong J, et al. Identification of a Tumor Microenvironment-Related Gene Signature Indicative of Disease Prognosis and Treatment Response in Colon Cancer. Oxid Med Cell Longev 2021;2021:6290261. [Crossref] [PubMed]
- Kim S, Kim A, Shin JY, et al. The tumor immune microenvironmental analysis of 2,033 transcriptomes across 7 cancer types. Sci Rep 2020;10:9536. [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- Yang H, Jin W, Liu H, et al. Immune-Related Prognostic Model in Colon Cancer: A Gene Expression-Based Study. Front Genet 2020;11:401. [Crossref] [PubMed]
- Chen ST, Lin YL, Huang MT, et al. CLEC5A is critical for dengue-virus-induced lethal disease. Nature 2008;453:672-6. [Crossref] [PubMed]
- Xiong W, Wang H, Lu L, et al. The macrophage C-type lectin receptor CLEC5A (MDL-1) expression is associated with early plaque progression and promotes macrophage survival. J Transl Med 2017;15:234. [Crossref] [PubMed]
- Chen PK, Hsieh SL, Lan JL, et al. Elevated Expression of C-Type Lectin Domain Family 5-Member A (CLEC5A) and Its Relation to Inflammatory Parameters and Disease Course in Adult-Onset Still's Disease. J Immunol Res 2020;2020:9473497. [Crossref] [PubMed]
- Elleisy N, Rohde S, Huth A, et al. Genetic association analysis of CLEC5A and CLEC7A gene single-nucleotide polymorphisms and Crohn's disease. World J Gastroenterol 2020;26:2194-202. [Crossref] [PubMed]
- Joyce-Shaikh B, Bigler ME, Chao CC, et al. Myeloid DAP12-associating lectin (MDL)-1 regulates synovial inflammation and bone erosion associated with autoimmune arthritis. J Exp Med 2010;207:579-89. [Crossref] [PubMed]
- Wortham BW, Eppert BL, Flury JL, et al. Cutting Edge: CLEC5A Mediates Macrophage Function and Chronic Obstructive Pulmonary Disease Pathologies. J Immunol 2016;196:3227-31. [Crossref] [PubMed]
- Chen R, Wu W, Chen SY, et al. A Pan-Cancer Analysis Reveals CLEC5A as a Biomarker for Cancer Immunity and Prognosis. Front Immunol 2022;13:831542. [Crossref] [PubMed]
- Xu F, Na L, Li Y, et al. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci 2020;10:54. [Crossref] [PubMed]
- Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin Cancer Biol 2019;59:125-32. [Crossref] [PubMed]
- Chen ST, Liu RS, Wu MF, et al. CLEC5A regulates Japanese encephalitis virus-induced neuroinflammation and lethality. PLoS Pathog 2012;8:e1002655. [Crossref] [PubMed]
- Yang MC, Wang CJ, Liao PC, et al. Hepatic stellate cells secretes type I collagen to trigger epithelial mesenchymal transition of hepatoma cells. Am J Cancer Res 2014;4:751-63. [PubMed]
- Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer 2014;14:430-9. [Crossref] [PubMed]
- Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 2014;15:786-801. [Crossref] [PubMed]
- Wang Y, Mei X, Song W, et al. LncRNA LINC00511 promotes COL1A1-mediated proliferation and metastasis by sponging miR-126-5p/miR-218-5p in lung adenocarcinoma. BMC Pulm Med 2022;22:272. [Crossref] [PubMed]
- Huang P, Li M, Tang Q, et al. Circ_0000523 regulates miR-1184/COL1A1/PI3K/Akt pathway to promote nasopharyngeal carcinoma progression. Apoptosis 2022;27:751-61. [Crossref] [PubMed]
(English Language Editor: J. Jones)


