
SOX9 knockout decreases stemness properties in colorectal cancer cells
Highlight box
Key findings
• SOX9’s absence affects the epithelial cell morphology and the pluripotency signature of the undifferentiated colorectal cell line HCT116.
What is known and what is new?
• SOX9 is an important transcription factor involved in the maintenance of stemness properties of normal colon cells.
• This is the first report showing evidence of SOX9 participation in CRC cell morphology. Also, we suggest two independent feedback loop effects between SOX9 and SOX2 as well as CD73.
What is the implication, and what should change now?
• More experiments are needed to deepen the feedback loop processes that we propose here, as well as the participation of SOX9 in cell morphology.
Introduction
Colorectal cancer (CRC) is one of the most relevant malignancies around the globe. It accounts for over 916,000 deaths being the 2nd most deadly cancer, affecting 1,931,590 individuals, positioning it in 3rd place of most common cancers worldwide in 2020 (1). Although more effective treatment procedures for CRC have been developed in recent years, the number of patients not responding to chemotherapy is still relevant (2). Besides, the patient relapse and metastasis have been associated with cell subpopulations present in solid tumors called tumor-initiating cells (TICs) or cancer stem cells (CSCs) (3-6).
CSCs share properties with mesenchymal stem cells (MSCs) such as self-renewal and differentiation. Specifically, CRC-CSCs present several surface markers such as CD144, CD166, CD29, ALDH1, LGR5 and CXCR4 but mainly CD133, CD24, and CD44 (7,8). Moreover, SOX2, OCT4, NANOG, and KLF4 transcription factors (TFs) are predominantly expressed in CSCs with a pivotal function in pluripotency maintenance (9-11).
Recently, CSCs and epithelial-mesenchymal transition (EMT) phenotype has been associated with the ability to invade and metastasize through the trans-differentiation of epithelial cells into a mesenchymal state (12,13). This event is mainly regulated by the activation of several genes, which turn off E-cadherin (CDH1) and turn on N-cadherin (CDH2) and Vimentin (VIM) (14,15). Also, this EMT activation is regulated by β-catenin (CTNNB1), the primary molecular effector of the Wnt signaling pathway (16).
Another essential regulator of the EMT phenotype is the SRY-box transcription factor 9 (SOX9), which also regulates organogenesis (17) and cell differentiation (18,19) in several tissues. SOX9 has a key role in the development of human cancer (20). Specifically, SOX9 has been associated to tumor progression, advanced tumor stage, and lower overall survival in CRC patients (21-24). While this participation of SOX9 on CRC has been demonstrated, its relationship with the stem phenotype has not been yet elucidated. Thus, we aim to assess the role of SOX9 in CRC-CSCs phenotype maintenance. We present this article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1163/rc).
Methods
Cell culture
The human CCR cell lines Caco-2, HT-29, HCT116 y SW-480 as well as the normal colon cell line CCD18-co, were obtained from American Type Culture Collection (ATCC). Culture conditions are those used by Lizárraga-Verdugo et al. (25). This study was conducted under the principles of the Declaration of Helsinki (as revised in 2013).
RNA isolation and reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA isolation and conditions for real-time PCR were developed following the methods of Lizárraga-Verdugo et al. (25). Real-time PCR was performed using TaqMan Gene Expression Assays (Applied Biosystems, CA, USA) and TaqMan Universal PCR Master Mix (Thermo Fisher Scientific, MA, USA) for SOX9 (Hs00165814_m1), OCT4 (Hs04260367_gH), KLF4 (Hs00358836_m1), SOX2 (Hs04234836_s1), NANOG (Hs02387400_g1), CTNNB1 (Hs00355045_m1), VIM (Hs00958111_m1), CDH1 (Hs01023895_m1), CDH2 (Hs00983056_m1) and ACTB (Hs01060665_g1) as endogenous control.
Flow cytometry immunophenotyping
Cultured cells were washed with phosphate-buffered saline (PBS) and resuspended in DMEM/F12 supplemented with 10% fetal bovine serum (FBS) at a density of 1×105 cells/100 µL. Single cells were stained using antibodies against CSCs; anti-human CD133-APC (Miltenyi Biotec, NRW, Germany), anti-human CD44-FITC (Miltenyi Biotec), and anti-human CD24-FITC (Miltenyi Biotec), and the Human MSC Analysis Kit (BD Biosciences, NJ, USA); CD90-FITC, CD105-PerCP-Cy5.5, and CD73-APC. Cells were incubated with the antibodies for 15 min at room temperature, washed with PBS, and analyzed using a BD Accuri C6 flow cytometer (BD Biosciences) and the software FlowJo v.10.
SOX9 knockout (KO) by CRISPR-Cas9
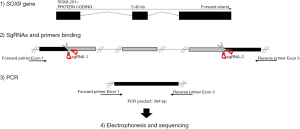
SOX9 KO was achieved using the CRISPR-Cas9 system. Single guide RNAs (sgRNA) for exon 1 and 3 of the SOX9 gene were designed using the Web application https://benchling.com/, obtaining the gene-specific sgRNA sequences 5'-CAGGAGAACACGTTCCCCAA-3' for exon 1 and 5'-ACGTCGCGGAAGTCGATAGG-3' for exon 3 (both synthesized by Integrated DNA Technologies, IDT, IA, USA). Cells (3×105) were grown on 6-well plates to perform a transfection of the CRISPR-cas9 system as follows: 26.9 µL Opti-MEM (Gibco, CA, USA), 2.6 µL Cas9 Plus Reagent (Invitrogen, CA, USA), 6,250 ng TrueCut Cas9 Protein v.2 (Invitrogen) and 100 pg of each sgRNAs, at the same time Lipofectamine CRISPRMAX Cas9 (Invitrogen) were diluted in 29.3 µL Opti-MEM. Both mixtures were incubated separately for 5 min at room temperature, then mixed and added to the cells. The cells were incubated with the mixture for 48 h and subsequently harvested to proceed with the cloning of single cells in a 96-well plate; when reaching sufficient cell density, a part of the cloned cells was taken for evaluation by PCR using the following primers for exon 1: forward 5'-CCCGCGTATGAATCTCCTG-3', and the reverse for exon 3 5'-TGCTTGGACATCCACACG-3' (both synthesized by IDT). This generated 384 bp PCR product was resolved by electrophoresis in 1.5% agarose gel at 100 V and analyzed by Sanger sequencing (Eton Bioscience, Inc., CA, USA) to confirm the SOX9-KO (Figure 1).
Immunofluorescence
For SOX9 localization 8×104 cells were grown in round-glass coverslips into a 24-well plate. Conditions and concentrations are those used by Lizárraga-Verdugo et al. (25). Finally, actin filaments were stained with 1:2,000 Phalloidin-iFluor 488 Reagent (Abcam, ab176753, Cambridge, UK) for 5 min, washed, and mounted with Fluoroshield with DAPI (Sigma-Aldrich, MA, USA). Visualization was obtained at a 40× magnification in a Leica LSCM (TCS SP8, Leica Microsystems, Wetzlar, Germany) and F-actin quantification was performed by phalloidin analysis of the image using the Leica software LAS X (v.3.5.6.21594) to obtain the mean values of relative fluorescence. Analyses were performed in triplicates from three different regions of interest (ROIs).
Sphere-formation, self-renewal, and differentiation assays
Cells were detached with 0.05% trypsin/EDTA (Gibco), and the resulting dissociated single cells were diluted to a density of 1×104 cells and placed into ultra-low attachment (ULA) 24-well plates with sphere formation medium (SFM), composed of serum-free medium DMEM/F12 supplemented with 20 ng/mL basic fibroblast growth factor (bFGF), 20 ng/mL epidermal growth factor (EGF) (both from ABM, BC, Canada) and 1× B27 supplement (Gibco). The cells were incubated at 37 ℃ with 5% CO2 for 14 days to let the formation of first-generation colonospheres.
To stimulate no CSCs differentiation or to evaluate self-renewal, first-generation spheroids were harvested and enzymatically dissociated with ACCUTASE (STEMCELL technologies, WA, USA) and gentle pipetting, then, 1×104 single cells, were re-plated and incubated again for 14 days, either in SFM, to examine the ability of colonospheres to form second-generation spheroids, or in DMEM/F12 supplemented with 15% FBS, to evaluate the loss of stemness maintaining ULA conditions (differentiation) by CD44, CD133 and CD24 evaluation using flow cytometer (previously mentioned). The number of spheres was assessed by counting under an inverted microscope using 10× objective. Also, for cell density analysis we performed measurements by using ImageJ software (https://imagej.nih.gov/ij/).
Statistical analysis
Three independent experiments in triplicate were performed and results were calculated as means ± standard deviation (SD) and analyzed using GraphPad Prism 8 software (MA, USA). Statistically significant differences between groups were assayed by a two-tailed t-test; multiple comparisons were tested by one-way analysis of variance (ANOVA) and Tukey’s post hoc test. A P<0.05 was considered significant.
Results
HCT116 exhibits more stemness phenotype than other CRC cell lines
To determine the colorectal cell line with more stem phenotype, we analyzed the expression of the pluripotency genes SOX2, KLF4, NANOG, and OCT4 by RT-qPCR in the CRC cell lines HCT116, HT-29, Caco-2, SW480 and the normal colon cell line CCD18-co. Caco-2 showed the highest levels of SOX2 in comparison to the control [fold change (FC =202.0, P<0.0001]; however, HCT116 was the cell line with the highest level of KLF4 (FC =11.1, P<0.001), NANOG (FC =29.6, P<0.001) and OCT4 (FC =35.5, P<0.001), but the third in expression of SOX2 (FC =16.8, P<0.0001) (Figure 2). To help guide our final selection, we also analyzed the surface markers CD44, CD133, and CD24 related to CRC-CSCs. HT-29 cells were almost all CD24+ cells (99.7%±0.1%), followed by SW-480, Caco-2 and HCT116, which showed more moderate positive levels (69.0%±16.1%, 67.1%±5.9% and 47.2%±2.06%, respectively). HCT116 had the highest levels of CD44+ cells (98.7%±1.0%) and the second-high population of CD133+ (90.9%±2.9%). Considering this, we settled for HCT116 for its high stem phenotype (Table 1).
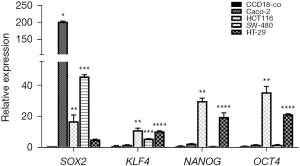
Table 1
| Cell line | % CD133 | % CD44 | % CD24 |
|---|---|---|---|
| HCT116 | 90.9±2.9 | 98.7±1.0 | 47.2±2.06 |
| HT-29 | 42.4±10.0 | 66.8±11.5 | 99.7±0.1 |
| Caco-2 | 97.8±1.1 | 14.3±2.7 | 67.1±5.9 |
| SW-480 | 0.4±0.2 | 81.6±8.1 | 69.0±16.1 |
| CCD18-co | 13.1±0.9 | 98.5±1.8 | 96.9±0.8 |
Absence of SOX9 induced morphological changes in HCT116
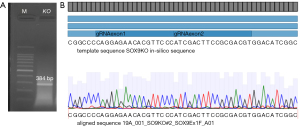
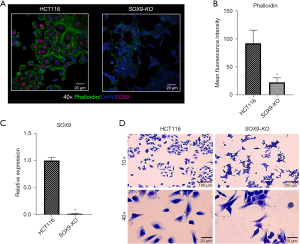
To assess the role of SOX9 gene in the stemness properties of the CRC HCT116 cell line, we performed a KO using the CRISPR-Cas9 system, obtaining one modified clone in which SOX9 protein expression was abolished. Our SOX9-KO clone had a deletion of 2,101 bp between exon 1 and 3, in both alleles (Figure 3A,3B). We then compared the expression of SOX9 at the protein (Figure 4A,4B) and mRNA (FC =0.01, P=0.0237) (Figure 4C) levels in both, the KO and parental cell lines, confirming that SOX9 was absent in SOX9-KO. Also, phalloidin levels were quantified showing a significant inhibition of this in SOX9-KO cells (P=0.0001) (Figure 4B). In addition, morphology analysis using Wright’s stain showed strikingly different morphology between SOX9-KO and its parental cell line; while parental HCT116 cells showed an expanded cytoplasm with a flatted, enlarged, and spread morphology, SOX9-KO cells displayed less cytoplasm with a spindle-shaped fibroblast-like morphology (Figure 4D). Despite the need for further studies, this morphological change could be explained by the change in actin filament reorganization, as observed by phalloidin stain (Figure 4A). These results suggest that SOX9 plays a role in the epithelial morphology of the HCT116 cell line.
KO of SOX9 in HCT116 affected its pluripotency signature, stem-cell markers, and EMT phenotype
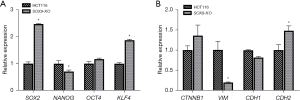
We evaluated the pluripotency genes SOX2, KLF4, NANOG, and OCT4 by RT-qPCR to elucidate the effect of SOX9 deletion on stemness properties, finding a significant overexpression of SOX2 (FC =2.5, P<0.0001) and KLF4 (FC =1.9, P<0.0001) in SOX9-KO in comparison with parental cell line HCT116, and a minimal non-significant rise of OCT4 (FC =1.2, P=0.0983) and NANOG downregulation (FC =0.7, P=0.0064) (Figure 5A). This indicates that SOX9 participates in the pluripotency signature. To expand our stem phenotype characterization, we measured MSCs surface markers such as CD90, CD105, and CD73 in both SOX9-KO and parental HCT116 line, finding a very low proportion of CD90 (0.9%±0.8% vs. 0.2%±0.2%, P=0.510), and CD105 (2.8%±0.5% vs. 0.7%±0.3%, P=0.052) positive cells; however, we found a significant inhibition of CD73 (43.3%±3.9% vs. 95.2%±2.9%, P=0.002), meaning that the expression of this marker could be affected by SOX9 absence. To evaluate if SOX9 absence affects EMT, we analyzed VIM, CDH1, CDH2, and CTNNB1 expression by RT-qPCR, finding a downregulation of VIM (FC =0.2, P=0.0017) in SOX9-KO cells in comparison with parental cells; in contrast, we observed an upregulation of CDH2 (FC =1.48, P=0.0363) (Figure 5B) in SOX9-KO cells. This suggests that the lack of SOX9 has a minimal impact on the EMT phenotype.
The absence of SOX9 inhibits tumorigenesis during the first- and second-sphere formation
To evaluate the effect of SOX9 deletion on tumorigenesis of CRC HCT116 line, we analyzed colonospheres generated in our first sphere-formation assay. The shape of the HCT116-spheres was spherical with regular and continuous contour, which can be described as compact tumor-packaging, meanwhile, SOX9-KO-spheres formed structures composed of rounded cells with weak neighboring cell-cell interactions (Figure 6A). SOX9-KO cells had a slightly higher tendency [P value = not significant (ns)] to form more spheres of their weaker and less compact characteristics (Figure 6B). Also, parental HCT116 cells formed larger spheres with diameters varying from 70 to 230 µm (Figure 6C), in comparison to SOX9-KO-spheroids which had a significantly lower cell density and size (40–160 µm) (P=0.0009) (Figure 6C).
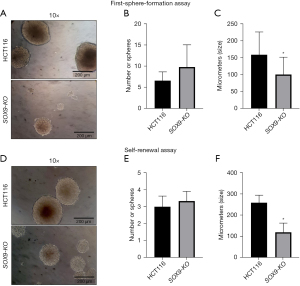
Next, we evaluated their self-renewal capability (second-colonospheres generation) finding similar observations to those of the first-sphere generation assay, in both the parental and SOX9-KO spheres (Figure 6D-6F). The above results suggest that SOX9-KO cells are inefficient in maintaining stable spheroids and triggering more spheres formation with less size. We hypothesize that this phenomenon is linked to morphological changes and its possible structural cytoskeleton behavior after SOX9 abolition.
SOX9 KO does not affect CSCs enrichment during sphere-formation assays
To evaluate the effect of SOX9 deletion on CSCs surface markers before and after first- and second-colonospheres formations, we analyzed the surface expression of CD133, CD44, and CD24 markers in parental and KO lines. During conventional cell culture (before spheres formation) we observed a significant inhibition of the initial basal levels on the positive population to CD44 from 98.7% in parental cells to 61.8% in SOX9-KO (P=0.042). In contrast, the population positivity for CD24 and CD133 did not change their proportions, showing ~90% and ~45% of cell positivity in both cell lines (Table 2).
Table 2
| Cell types | % CD133 | % CD44 | % CD24 |
|---|---|---|---|
| Basal | |||
| HCT116 | 90.9±2.9 | 98.7±1.0 | 47.2±2.06 |
| SOX9-KO | 87.8±3.1 | 61.8±14.4a | 45.1±10.3 |
| First-generation-spheres | |||
| HCT116 | 90.5±1.3 | 96.0±6.0 | 67.1±11.9 |
| SOX9-KO | 79.7±5.4 | 88.0±12.4 | 75.0±12.2b |
| Second-generation-spheres | |||
| HCT116 | 92.8±1.7 | 98.7±1.01 | 79.4±8.9 |
| SOX9-KO | 88.8±2.7 | 97.3±0.9 | 83.6±18.7 |
| Differentiation assay | |||
| HCT116 | 11.9±0.0c | 82.8±0.2 | 24.8±2.4c |
| SOX9-KO | 17.2±0.0c | 71.3±3.8c | 22.8±3.4c |
a, P<0.05 comparing HCT116 vs. SOX9-KO basal surface marker proportions; b, P<0.05 comparing HCT116 and SOX9-KO from first-generation-spheres vs. basal marker proportions; c, P<0.05 comparing HCT116 and SOX9-KO from differentiation vs. first-generation-spheres marker proportions. The data are shown as mean ± standard deviation. CSC, cancer stem cell; SOX9, SRY-box transcription factor 9; KO, knockout.
During the first-colonosphere generation assay, we found similar proportions of CD44 and CD24 in both groups, while CD133 population was lower (90.5%, P=0.052) in SOX9-KO (79.7%) as compared to HCT116 (Table 2). Similarly, after the sphere self-renewal assay (second-colonosphere formation) no changes were found between SOX9-KO and HCT116 in any marker proportion (Table 2).
When comparing CSCs marker levels of first-generation-colonospheres, in KO and parental cells, against their respective initial basal levels, SOX9-KO line showed no significant changes for CD133 (from 87.8% to 79.7%, P=0.240) and CD44 (from 61.8% to 88.0%, P=0.220), and a significant increase on CD24 from 45.1% to 75.0% (P=0.015). No changes were found for HCT116 in any marker (Table 2). Additionally, a comparison between first- and second-generation-colonospheres, showed no differences in any marker for both lines.
SOX9 does not associate the loss of CSCS markers, in both the KO and its parental line
To investigate the role of SOX9 on CSCs differentiation, we promoted differentiation to no CSCs of dissociated first-colonospheres by adding 15% FBS and keeping ULA conditions. Differentiated cultures were then analyzed for CSCs differentiation surface markers. Both, HCT116 and SOX9-KO cells diminished their percentages of CD133 (90.5%±1.3% vs. 11.9%±0.0%, P=0.0001; 79.7%±5.4% vs. 17.2%±0.0%, P=0.0002, respectively) and CD24 (67.1%±11.9% vs. 24.8%±2.4%, P=0.023; 75.0%±12.2% vs. 22.8%±3.4%, P=0.010, respectively) positive cells (Table 2). Considering that SOX9-KO cells observed the same behavior as its parental line, with a fall on CD44 (88.0%±12.4% vs. 71.3%±3.8%, P=0.006) positive cells (Table 2), we concluded that SOX9 does not seem to participate in the loss of HCT116 CSCs surface markers.
Discussion
SOX9 is often overexpressed in solid cancers, including CRC (26), but its role in CSCs maintenance is unclear. In this work, we reported a clear association between SOX9 and CSCs characteristics in a CRC line.
The co-expression of SOX2, OCT4, KLF4, and NANOG is essential to promote stemness by upregulating genes involved in self-renewal and pluripotency and suppressing genes implicated in cell differentiation (27). In this regard, we measured the expression of these TFs in CRC cell lines, our finding showed that HCT116 exhibits the strongest stem phenotype of the cell lines analyzed (Figure 2). In agreement with our results (Table 1), it has been reported that HCT116 exhibits a low CD24+ accompanied by a moderate (28) or high CD44/CD133 population (29). Previously, our research group found that SOX9 is overexpressed in HCT116 compared to other CRC cell lines (26) and was, therefore, the cell line of choice to KO the SOX9 gene (SOX9-KO).
As an effect, we observed that cell morphology, actin-filaments reorganization, and f-actin were seriously affected in SOX9-KO (Figure 4A,4D). A previous study showed that ectopic SOX9 expression turns on a flatter morphological cell appearance as compared with the control in prostate cancer cell lines; the above is due to overexpression of genes involved in cell adhesion and shape (30).
An increased expression of SOX2 and KLF4 and decreased NANOG were observed in our SOX9-KO cells (Figure 5A). Interestingly; the reduction of SOX9 expression resulted in a feedback loop effect with SOX2, due to their reciprocal binding to their promoter regions in breast cancer cell lines (31). While the above findings derived from a partial silencing of SOX9, our findings resulted from a KO, suggesting a possible compensatory process due to a total absence of SOX9 and resulting in a SOX2 overexpression. In addition, ectopic SOX9 expression downregulates KLF4 expression (32). Interestingly, in normal colon, KLF4 is highly expressed in differentiated cells (33) and poorly expressed in the crypts where the undifferentiated cells are found; meanwhile, SOX9 has the opposite pattern (33). Together, these results support the regulatory role of SOX9 on pluripotency genes related to the stemness phenotype of the undifferentiated CRC cell line HCT116.
In the MSCs markers context, reports have shown that CD73 promotes SOX9 transcription, and protects to SOX9 from degradation in hepatocellular carcinoma (34). Interestingly, as the sudden decrease in CD73+ cells in our SOX9-KO we propose a possible positive regulation of CD73 by SOX9. However, more information is needed to understand this mechanism.
SOX9 is an important promoter of EMT phenotype (35), however, we observed a maintenance of this phenotype, even with the absence of SOX9. Interestingly, CD73 has been related to EMT since it regulates at transcriptional levels essential EMT driver genes such as TWIST1, SNAI1, and ZEB1 (36). As we showed before, we have a partial presence of this marker, showing that the EMT phenotype could be gained in SOX9 absence. Further experiments must confirm the above to elucidate their specific role.
Interestingly, we obtained a tendency to a larger number of small size spheres in the SOX9-KO cell line, in both first- and second generation-spheres (Figure 6), the above possibly due to by the morphology change present by SOX9 abolition which result in labile spheres.
We found an initial effect on CD44 basal levels in SOX9-KO line (Table 2), but only before the first and second sphere-formation assays; these results could be due to the effect of components of the sphere medium such as the B27, which allows tumor-spheres growth in the absence of FBS, promoting CSCs enrichment (37); also, the presence of FGF and EGF factors which promote CSCs enrichment (38-40). Interestingly, we found an increase of CD24 after first-generation-spheres of SOX9-KO cells, in comparison with its own initial basal level.
Finally, it has been demonstrated that the presence of FBS stimulates the loss of the surface markers related to CSCs; such as CD24, which declined in CSCs derived from hepatocellular carcinoma tissues, promoting the differentiation and recovering the phenotype of the cells before CSCs enrichment (41). This suggests that FBS stimulates the loss of CSCs by cell differentiation, which was observed in our results. We must highlight here the need of our research field to harmonize culture conditions for cell lines, amenable with proper in vivo cell micro-environments. The use of xenogeneic factors or the unsuspected presence of endotoxins in our culture media must be carefully considered giving preference to human additives rather than bovine.
Conclusions
In conclusion, SOX9 absence affects the epithelial morphology and the pluripotency signature of the HCT116, an undifferentiated colorectal cell line. We found an increase in SOX2 expression as a compensatory mechanism by SOX9 lack, and the increase of KLF4 could be associated with an early differentiation process. Also, SOX9 is associated with the presence of CD44 and CD73 positive populations, being the last one a putative feedback loop effect between CD73 and SOX9. Interestingly, the EMT phenotype is still occurring in SOX9-KO cells, possibly by compensatory mechanisms. The lower size and compaction of the spheres in SOX9-KO could be influenced by the morphology change. Collectively, these results provide evidence that SOX9 promotes the maintenance of stemness properties in CRC-CSCs.
Acknowledgments
Funding: This research was funded by Consejo Nacional de Ciencia y Tecnologia (CONACYT), Mexico, grants Programa Presupuestario F003 #51207/2020 and 290311.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1163/rc
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1163/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1163/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted under the principles of the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- WHO. Cancer Fact sheet 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer
- Van Cutsem E, Cervantes A, Adam R, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016;27:1386-422. [Crossref] [PubMed]
- Hu Y, Yan C, Mu L, et al. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS One 2015;10:e0125625. [Crossref] [PubMed]
- Abdul Khalek FJ, Gallicano GI, Mishra L. Colon cancer stem cells. Gastrointest Cancer Res 2010;S16-23. [PubMed]
- Ayob AZ, Ramasamy TS. Cancer stem cells as key drivers of tumour progression. J Biomed Sci 2018;25:20. [Crossref] [PubMed]
- Huang R, Rofstad EK. Cancer stem cells (CSCs), cervical CSCs and targeted therapies. Oncotarget 2017;8:35351-67. [Crossref] [PubMed]
- Fedyanin M, Anna P, Elizaveta P, et al. Role of Stem Cells in Colorectal Cancer Progression and Prognostic and Predictive Characteristics of Stem Cell Markers in Colorectal Cancer. Curr Stem Cell Res Ther 2017;12:19-30. [Crossref] [PubMed]
- Stoian M, Stoica V, Radulian G. Stem cells and colorectal carcinogenesis. J Med Life 2016;9:6-11. [PubMed]
- Dai X, Ge J, Wang X, et al. OCT4 regulates epithelial-mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncol Rep 2013;29:155-60. [Crossref] [PubMed]
- Wen K, Fu Z, Wu X, et al. Oct-4 is required for an antiapoptotic behavior of chemoresistant colorectal cancer cells enriched for cancer stem cells: effects associated with STAT3/Survivin. Cancer Lett 2013;333:56-65. [Crossref] [PubMed]
- Hu J, Li J, Yue X, et al. Expression of the cancer stem cell markers ABCG2 and OCT-4 in right-sided colon cancer predicts recurrence and poor outcomes. Oncotarget 2017;8:28463-70. [Crossref] [PubMed]
- Wang SS, Jiang J, Liang XH, et al. Links between cancer stem cells and epithelial-mesenchymal transition. Onco Targets Ther 2015;8:2973-80. [PubMed]
- Vand-Rajabpour F, Noormohammadpour P, Ahmadifard MR, et al. Upregulation of SNAI2 and SOX9 mRNA versus downregulation of eight other EMT/stemness related genes in basal cell carcinoma. Br J Dermatol 2019;181:1065-6. [Crossref] [PubMed]
- Vu T, Datta PK. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers (Basel) 2017;9:171. [Crossref] [PubMed]
- Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol 2017;11:28-39. [Crossref] [PubMed]
- Kim WK, Kwon Y, Jang M, et al. β-catenin activation down-regulates cell-cell junction-related genes and induces epithelial-to-mesenchymal transition in colorectal cancers. Sci Rep 2019;9:18440. [Crossref] [PubMed]
- Hong Y, Chen W, Du X, et al. Upregulation of sex-determining region Y-box 9 (SOX9) promotes cell proliferation and tumorigenicity in esophageal squamous cell carcinoma. Oncotarget 2015;6:31241-54. [Crossref] [PubMed]
- Mori-Akiyama Y, van den Born M, van Es JH, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 2007;133:539-46. [Crossref] [PubMed]
- Lefebvre V, Dvir-Ginzberg M. SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect Tissue Res 2017;58:2-14. [Crossref] [PubMed]
- Aguilar-Medina M, Avendaño-Félix M, Lizárraga-Verdugo E, et al. SOX9 Stem-Cell Factor: Clinical and Functional Relevance in Cancer. J Oncol 2019;2019:6754040. [Crossref] [PubMed]
- Song W, Kwon GY, Kim JH, et al. Immunohistochemical staining of ERG and SOX9 as potential biomarkers of docetaxel response in patients with metastatic castration-resistant prostate cancer. Oncotarget 2016;7:83735-43. [Crossref] [PubMed]
- Matheu A, Collado M, Wise C, et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res 2012;72:1301-15. [Crossref] [PubMed]
- Bruun J, Kolberg M, Nesland JM, et al. Prognostic Significance of β-Catenin, E-Cadherin, and SOX9 in Colorectal Cancer: Results from a Large Population-Representative Series. Front Oncol 2014;4:118. [Crossref] [PubMed]
- Shen Z, Deng H, Fang Y, et al. Identification of the interplay between SOX9 and S100P in the metastasis and invasion of colon carcinoma. Oncotarget 2015;6:20672-84. [Crossref] [PubMed]
- Lizárraga-Verdugo E, Ruiz-García E, López-Camarillo C, et al. Cell Survival Is Regulated via SOX9/BCL2L1 Axis in HCT-116 Colorectal Cancer Cell Line. J Oncol 2020;2020:5701527. [Crossref] [PubMed]
- Lizárraga-Verdugo E, Carmona TG, Ramos-Payan R, et al. SOX9 is associated with advanced T-stages of clinical stage II colon cancer in young Mexican patients. Oncol Lett 2021;22:497. [Crossref] [PubMed]
- Hadjimichael C, Chanoumidou K, Papadopoulou N, et al. Common stemness regulators of embryonic and cancer stem cells. World J Stem Cells 2015;7:1150-84. [Crossref] [PubMed]
- Muraro MG, Mele V, Däster S, et al. CD133+, CD166+CD44+, and CD24+CD44+ phenotypes fail to reliably identify cell populations with cancer stem cell functional features in established human colorectal cancer cell lines. Stem Cells Transl Med 2012;1:592-603. [Crossref] [PubMed]
- Ke J, Wu X, Wu X, et al. A subpopulation of CD24+ cells in colon cancer cell lines possess stem cell characteristics. Neoplasma 2012;59:282-8. [Crossref] [PubMed]
- Francis JC, Capper A, Ning J, et al. SOX9 is a driver of aggressive prostate cancer by promoting invasion, cell fate and cytoskeleton alterations and epithelial to mesenchymal transition. Oncotarget 2018;9:7604-15. [Crossref] [PubMed]
- Domenici G, Aurrekoetxea-Rodríguez I, Simões BM, et al. A Sox2-Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene 2019;38:3151-69. [Crossref] [PubMed]
- Flandez M, Guilmeau S, Blache P, et al. KLF4 regulation in intestinal epithelial cell maturation. Exp Cell Res 2008;314:3712-23. [Crossref] [PubMed]
- Ghaleb AM, McConnell BB, Kaestner KH, et al. Altered intestinal epithelial homeostasis in mice with intestine-specific deletion of the Krüppel-like factor 4 gene. Dev Biol 2011;349:310-20. [Crossref] [PubMed]
- Ma XL, Hu B, Tang WG, et al. CD73 sustained cancer-stem-cell traits by promoting SOX9 expression and stability in hepatocellular carcinoma. J Hematol Oncol 2020;13:11. [Crossref] [PubMed]
- Choi BJ, Park SA, Lee SY, et al. Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9. Sci Rep 2017;7:15918. [Crossref] [PubMed]
- Lupia M, Angiolini F, Bertalot G, et al. CD73 Regulates Stemness and Epithelial-Mesenchymal Transition in Ovarian Cancer-Initiating Cells. Stem Cell Reports 2018;10:1412-25. [Crossref] [PubMed]
- Gu Y, Fu J, Lo PK, et al. The effect of B27 supplement on promoting in vitro propagation of Her2/neu-transformed mammary tumorspheres. J Biotech Res 2011;3:7-18.
- Shimabukuro Y, Terashima H, Takedachi M, et al. Fibroblast growth factor-2 stimulates directed migration of periodontal ligament cells via PI3K/AKT signaling and CD44/hyaluronan interaction. J Cell Physiol 2011;226:809-21. [Crossref] [PubMed]
- Maehara O, Suda G, Natsuizaka M, et al. Fibroblast growth factor-2-mediated FGFR/Erk signaling supports maintenance of cancer stem-like cells in esophageal squamous cell carcinoma. Carcinogenesis 2017;38:1073-83. [Crossref] [PubMed]
- Feng Y, Dai X, Li X, et al. EGF signalling pathway regulates colon cancer stem cell proliferation and apoptosis. Cell Prolif 2012;45:413-9. [Crossref] [PubMed]
- Lee TK, Castilho A, Cheung VC, et al. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell 2011;9:50-63. [Crossref] [PubMed]


