
Esophageal cancer stem cells reduce hypoxia-induced apoptosis by inhibiting the GRP78-perk-eIF2α-ATF4-CHOP pathway in vitro
Highlight box
Key findings
• Based on the GRP78-PERK-eIF2α-ATF4-CHOP signaling pathway, esophageal CSCs can resist hypoxia-induced apoptosis by inhibiting the induction of CHOP overexpression by hypoxia.
What is known and what is new?
• Researches has found the presence of CSCs in esophageal cancer, which can resist hypoxia-induced cell apoptosis. Based on this characteristic, CSCs endow esophageal cancer with drug resistance and high recurrence rate during treatment.
• This study found that the molecular mechanism of resistance of esophageal CSCs to hypoxia lies in the inhibition of CHOP mediated ERS apoptosis pathway by CD44+CD24− tumor stem cells in a hypoxic environment and further inhibit the activation of the mitochondrial apoptosis pathway.
What is the implication, and what should change now?
• This study will provide a new perspective for explaining the resistance of esophageal CSCs to hypoxia-induced apoptosis, and new ideas for the treatment of esophageal cancer.
Introduction
Esophageal cancer (EC) is one of the common malignant tumors (1). Research has found that factors influencing the occurrence and development of EC include smoking, alcohol consumption, diet, infection, and genetics (2). The pathogenesis of EC involves abnormalities at multiple molecular levels, including DNA, messenger RNA (mRNA), microRNA (miRNA), long non-coding RNA (lncRNA), and proteins. Previous studies have found that lncRNA HAGLROS may play a role in the progression of EC by regulating the miR-206/NOTCH3 signaling axis in EC, and it may become a new target for the diagnosis and treatment of EC (3). However, there are currently no effective drugs and strategies for EC based on molecular targets. Meanwhile, EC exhibits strong resistance to conventional chemotherapy drugs. The mechanisms mediating drug resistance include apoptosis inhibition protein family, PI3K/AKT/mTOR pathway, NF-κB pathway, p38 and p53 signaling pathways, tumor stem cells, tumor microenvironment (TME) transformation, invasion and metastasis, and metabolism (4). Among these, the resistance of EC conferred by tumor stem cells has been confirmed by multiple studies and significantly impacts the efficacy of conventional drug treatments.
EC cells have been found to contain tumor cells with stem cell properties (5,6), called cancer stem cells (CSCs), which are characterized by carcinogenicity, multiple differentiation, self-renewal, and drug resistance (7). These characteristics were shown to be the main reason for strong drug resistance, poor prognosis, and recurrence of EC (8-10). Although recent studies have found that hypoxia could induce apoptosis of tumor cells (11), CSCs have a strong survival ability in hypoxic environments (12). Although inhibition of tumor angiogenesis can lead to the apoptosis of most tumor cells in antiangiogenic tumor therapy (13-15), CSCs can survive in this hypoxic environment and promote tumor recurrence (16-18). Therefore, it is of great significance to explore the anti-hypoxic induction mechanism of CSCs to treat EC.
To date, it has been found that there are three main pathways leading to apoptosis, including the mitochondrial apoptosis pathway, Fas receptor-mediated apoptosis pathway, and ERS mediated apoptosis pathway. The activation of the above three apoptosis pathways can finally regulate apoptosis by stimulating the cleavage and activation of caspase 3, caspase 6, and caspase 7 proteins.
Studies have found that continuous hypoxia can activate endoplasmic reticulum stress (ERS) (19,20). On the one hand, ERS can keep cells alive by activating unfolded protein response (UPR) signaling pathways within the acceptable range (21). On the other hand, excessive ERS can cause apoptosis (19). Prolonged hypoxia has been shown to lead to a large accumulation of misfolded proteins, causing excessive ERS and inducing apoptosis (22).
The GRP78-PERK-eIF2α-ATF4 signaling pathway plays an important role in UPR and apoptosis induced by ERS (23,24). In the absence of ERS, PERK exists in an inactive form by binding to GRP78/Bip. During ERS, PERK dissociates from GRP78/Bip and becomes activated. In UPR, activated PERK phosphorylates eIF2α, resulting in the inactivation of eIF2α, which terminates the synthesis of most proteins in the cell and regulates the downstream cell cycle. In ERS-induced apoptosis, the PERK-eIF2α pathway activates ATF4 and causes CHOP expression. Upregulated CHOP-induced apoptosis is achieved by the following means: (I) CHOP leads to growth arrest and DNA damage-inducing protein 34, accelerates p-eIF2α dephosphorylation, and a large number of proteins are translated, increasing cell load and inducing apoptosis (25). (II) In normal cells, the anti-apoptotic Bcl-2 gene maintains a relative balance with the pro-apoptotic protein Bax. Excessive CHOP will inhibit the transcription of Bcl-2, resulting in a relative excess of Bax, and the increased free Bax is transported to mitochondria. Inducing mitochondrial pathway apoptosis (26). (III) CHOP upregulates the expression of endoplasmic reticulum oxidase 1α, which produces a multitude of reactive oxygen species (ROS) and calcium ions during disulfide bond formation. Free calcium ions enter mitochondria and depolarize the inner membrane to generate ROS, during which adenosine triphosphate (ATP) is provided by mitochondria. Therefore, ERS induces oxidative stress and disrupts mitochondrial function, leading to endogenous apoptosis. (IV) CHOP promotes the expression of Tribbles homolog 3 and subsequently inhibits the phosphorylation of anti-apoptotic PKB/AKT, reduces PKB activity, and induces apoptosis (27).
It has been found that CHOP overexpression can promote EC cell apoptosis (28), but whether esophageal CSCs can resist hypoxia-induced apoptosis by inhibiting the induction of GRP78-PERK-eIF2α-ATF4-CHOP signaling pathway by hypoxia has not been studied. In this study, CD44+CD24− cells in EC9706 cells were screened as tumor stem cell subtypes. EC9706 cells and CD44+CD24− cells were treated with hypoxia, respectively. To explore the resistance of esophageal CSCs to hypoxia, the effects of hypoxia on apoptosis were detected. To clarify which apoptosis pathway esophageal CSCs resist hypoxia-induced cell apoptosis through, the effects of hypoxia on the expression of apoptosis-related proteins and the apoptosis pathway were detected. To clarify the mechanisms of CD44+CD24− cells resistance to hypoxia-induced apoptosis is achieved by inhibiting the activation of ERS pathway, silenced CHOP and PERK cell lines of EC9706 cells and overexpressed CHOP and PERK cell lines of CD44+CD24− cells were constructed. After hypoxia intervention, the effects of hypoxia on apoptosis, cell cycle, mitochondrial membrane potential and the expressions of apoptosis-related proteins Bax, Bcl-2, GADD34, Ero1-Lα, and TRIB3, and mitochondrial apoptosis-related proteins Bax, Bcl-2, Apaf, cytochrome C (Cyt C), and caspase 9 in the downstream of CHOP under hypoxia were detected. In summary, this study aimed to explore the molecular mechanism of esophageal CSCs inhibiting hypoxia-induced apoptosis and provide a theoretical basis for the prevention and treatment of EC and the development of targeted drugs. We present this article in accordance with the MDAR reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-462/rc).
Methods
Materials and reagents
The EC cell line EC9706 was purchased from Shanghai Yuchi Biotechnology Co., Ltd. (Shanghai, China; accession number: SC0152). CD44+CD24− cell line was screened as tumor stem cell subtypes by fluorescence-activated cell sorting (FACS) from EC9706 cell line.
High glucose-Dulbecco’s modified Eagle medium (H-DMEM; Gibco, Carlsbad, CA, USA); DMEM/F12 medium (Gibco); fetal bovine serum (FBS; Sijiqing Company, Hangzhou, China); 0.25% trypsin (T1300; Solarbio Co., Beijing, China); 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; M8180; Solarbio Co.); JC-1 (MAK160; Sigma-Aldrich, Waltham, MA, USA); annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (CA1020; Solarbio Co.); cycle detection kit (CA1510; Solarbio Co.); Cyt C releasing apoptosis assay kit (Ab65311; Abcam, Cambridge, MA, USA); radioimmunoprecipitation assay (RIPA) lysis buffer (P0013C; Beyotime Co., Shanghai, China); horseradish peroxidase (HRP)-labeled goat anti-rabbit secondary antibody (A0208; Beyotime Co.); real-time fluorescence quantitative polymerase chain reaction (qPCR) kit (RR820A; Bao Bioengineering Co., Ltd., Dalian, China); TRIeasyTM total RNA extraction reagent kit (9109; Bao Bioengineering Co., Ltd.); HyperScript III RT SuperMix for qPCR with genomic DNA (gDNA) Remover Kit [reverse transcription polymerase chain reaction (RT-PCR); RR047A; Bao Bioengineering Co., Ltd.]; rabbit anti-SOX2 polyclonal antibody (AB92494; Abcam); rabbit anti-OCT4 polyclonal antibody (AB200834; Abcam); rabbit anti-caspase 3 polyclonal antibody (AB13847; Abcam); rabbit anti-caspase 6 monoclonal antibody (AB185645; Abcam); rabbit anti-caspase 7 monoclonal antibody (AB255818; Abcam); phosphatidylethanolamine (PE)-conjugated anti-CD44 mouse monoclonal antibody (AB269300; Abcam); FITC-conjugated anti-CD24 mouse monoclonal antibody (AB30350; Abcam); HRP-labeled anti-rabbit secondary antibody (AB6721; Abcam); rabbit anti-caspase 9 monoclonal antibody (AB32539; Abcam); rabbit anti-Apaf-1 monoclonal antibody (AB234436; Abcam); rabbit anti-Bax monoclonal antibody (ab32503; Abcam); rabbit anti-Bcl-2 monoclonal antibody (ab32124; Abcam); rabbit anti-Cyt C monoclonal antibody (AB133504; Abcam); rabbit anti-VDAC1 monoclonal antibody (AB15895; Abcam); rabbit anti-Caspase 12 polyclonal antibody (ab62484; Abcam); rabbit anti-CHOP monoclonal antibody (ab11419; Abcam); rabbit anti-GRP78 monoclonal antibody (ab108615; Abcam); rabbit anti-JNK monoclonal antibody (ab199380; Abcam); rabbit anti-Ero1-Lα monoclonal antibody (ab177156; Abcam); rabbit anti-GADD34 monoclonal antibody (ab236516; Abcam); rabbit anti-TRIB3 monoclonal antibody (ab137526; Abcam); rabbit anti-ATF4 monoclonal antibody (ab270980; Abcam); rabbit anti-eIF2α monoclonal antibody (ab169528; Abcam); rabbit anti-p-eIF2α monoclonal antibody (YP0093; ImmunoWay Biotechnology, Plano, TX, USA); rabbit anti-PERK monoclonal antibody (ab229912; Abcam).
Study methods
Culture of EC9706 and CD44+CD24− cell lines
EC9706 cells were cultured in H-DMEM containing 10% FBS in a humidified atmosphere of 95% air, 5% CO2 at 37 ℃. CD44+CD24− cells were cultured in serum-free DMEM/F12 medium (containing 2% B27, 20 ng/mL EGF, 20 ng/mL bFGF, 10 µg/mL insulin, 0.5 µg/mL hydrocortisone, 0.4% bovine serum aluminum, and 2 mM L-glutamine) in a humidified atmosphere of 95% air, 5% CO2 at 37 ℃. When they reached 80% confluence, the cells were digested and subcultured with 0.25% trypsin in the ratio of 1:3.
Screening of esophageal CSCs characterized by CD44+CD24−
- Enrichment of stem cells by microsphere culture: after the cells had grown to a confluence of about 80%, 0.25% trypsin and serum-free DMEM/F12 medium [containing 2% B27, 20 ng/mL epidermal growth factor (EGF), 20 ng/mL basic fibroblast growth factor (bFGF), 10 µg/mL insulin, 0.5 µg/mL hydrocortisone, 0.4% bovine serum aluminum, and 2 mM L-glutamine] were prepared into a 1×103/mL single-cell suspension and was used for suspension culture. Then, 1 mL DMEM/F12 medium was added every 2 days for continuous culture for 14 days. After the cells had large clonal spheres, they were separated into single-cell suspension by mechanical blowing method, subcultured, and expanded to 10 bottles. The suspended cells were collected by centrifugation at 3,000 rpm for 5 minutes, then washed with phosphate-buffered saline [PBS; PBS containing 0.5% bovine serum albumin (BSA) and 2 mM ethylenediamine tetraacetic acid (EDTA) (PBS-BE)] for 2 times, and then washed with 1×107/mL density was resuspended in PBS-BE.
- Separation of CD44+/CD24− tumor stem cells by magnetic beads: 0.5 mL (about 5 mL) of EC9706 cell single-cell suspension obtained from suspension culture in (1×106) was transferred to a 5 mL centrifuge tube and 20 µL of human CD24 biotinylated antibody was added, followed by gentle mixing and incubation at 4 ℃ for 30 minutes. After incubation, 3 mL of precooled PBS-BE was added, followed by centrifugation at 300 g and washing for 10 minutes. The cells were then resuspended with 0.5 mL PBS-BE and 50 mL/µL streptavidin ferrofluid was added. After mixing, the cells were incubated at 4 ℃ for 30 minutes. After incubation, 3 mL of precooled PBS-BE was added and the cells were centrifuged at 300 g for 10 minutes, washed, the supernatant was discarded, 3 mL PBS-BE was added to resuspend cells. The resuspended solution was transferred to a new 5 mL centrifuge tube and placed in MagCellectTM Magnet (R & D Systems, Minneapolis, MN, USA), incubated at room temperature for 8 minutes. While keeping the centrifuge tube in the magnetic field, all suspensions were carefully absorbed with a sterile pasteurized pipette, transferred to a new 5 mL centrifuge tube, and the original centrifuge tube was discarded. After repeating this process 6 times, CD24+ cells were removed entirely and CD24low/− cells were obtained. CD24low/− cells were concentrated by centrifugation at 300 g for 10 minutes, resuspended with 0.5 mL PBS-BE, transferred to a 15 mL centrifuge tube, and 2 µL of human CD44 biotinylated antibody was added with 10 mL PBS-BE, followed by gentle thorough mixing, and incubation at 4 ℃ for 30 minutes. After incubation, 3 mL of precooled PBS-BE was added, centrifuged at 300 g for 10 minutes, and then washed. The cells were then resuspended with 0.5 mL PBS-PE and 50 mL 10 µL streptavidin ferrofluid was added. After mixing, the cells were incubated at 4 ℃ for 30 minutes, followed by the addition of 3 mL precooled PBS-BE, centrifugation at 300 g for 10 minutes, washing, disposal of the supernatant, and the addition of 3 mL PBS-BE to resuspend cells. The resuspended solution was then transferred to a new 5 mL centrifuge tube and placed it in MagCellectTM Magnet, incubated at room temperature for 8 minutes, and keeping the centrifuge tube in the magnetic field, all suspensions were carefully absorbed with a sterile pasteurized pipette and discarded. The centrifuge tube containing selected CD44high/+ cells from the magnetic field was then taken out, and resuspended with 3 mL of precooled PBS-BE cells. CD44+/CD24− cells were then obtained after repeating this process 6 times. Finally, the obtained CD44+/CD24− cells were resuspended with culture medium and cultured. The separation efficiency was measured by flow cytometry.
- The proportion of CD44+/CD24− cells sorted by magnetic beads was detected by flow cytometry: 0.5 mL of single-cell suspension of EC9706 cells in normal culture and 0.5 mL of CD44+/CD24− cells sorted by magnetic beads (about 5×105 cells) were added with 10 µL FITC-conjugated anti-CD24 mouse monoclonal antibody and 10 µL of the PE-conjugated anti-CD44 mouse monoclonal antibody, incubated at 4 ℃ for 30 minutes, centrifuged and washed twice with PBS, and added 400 µL PBS to resuspend cells. The proportion of CD44+/CD24− cells in unselected cells and sorted cells was detected by FACS.
Hypoxia treatment
EC9706 cells and CD44+/CD24− cells were cultured in a 0.1% O2 hypoxia incubator for 24 hours with 10% FBS-supplemented H-DMEM and serum-free DMEM/F12 treated by ultrasonic deaeration. The corresponding cells were cultured in a normoxic environment as normal control.
Construction of EC9706 cell line with low expression of CHOP and PERK genes
The recombinant lentivirus vectors for overexpressing or silencing the CHOP and PERK genes was constructed by Shanghai GeneChem Co., Ltd. (Shanghai, China). The brief steps were as follows.
The mRNA sequences of CHOP (NM_001195055) and PERK (NM_004836.7) were obtained from GeneBank (Shenzhen, China), and the short hairpin RNA (shRNA) sequences were designed as follows: CHOP shRNA A: 5'-CCGGTGGAACGCTCAAGGAAATCGCTGCTTGAGCCGTTTTTTTTTG-3', B: 5'-AATTCAAAAATGAACGGCTCAGCAGGAAATCGCTGCTTGCTTTTTTTTCA-3'; PERK shRNA A: 5'-CCGGCTCAAATTTCCACATTATTTCGAG AAATATGGGAAATTTGAGTTTG-3', B: 5'-AATTCAAAACTCAAATTTCCACATTATTTCGAGAAATATGGTGGAAATTTGAG-3', and TTCTCCGAACGACGTCACGT as negative control, the above sequences were synthesized respectively. The synthesized primers were dissolved in annealing buffer in pairs, placed in a 90 ℃ water bath for 15 minutes, and naturally cooled to room temperature, The DNA of double-stranded shRNA was obtained. The double enzyme digestion linearized GV493 vector was connected with annealed double-stranded DNA through T4 DNA ligase and connected at 16 ℃ for 2 hours. The recombinant product was transformed into DH5α, cultured on the screening plate, the monoclonal antibodies on the plate were selected for PCR identification, the positive clones were sequenced and the results analyzed to obtain the correct clone, expand the culture and extraction, and obtain high-purity plasmids. Subsequently, the 20 µg obtained vector plasmid, 15 µg pHelper 1.0 vector plasmid, and 10 µg pHelper 2.0 vector plasmid were mixed and slowly added to the culture medium of 293T cells after adding transfection reagent, mixed evenly, and cultured in the cell incubator at 37 ℃ and 5% CO2. After 72 hours, the supernatant was collected and ultracentrifuged to obtain the packaged lentivirus vector for lentivirus quality detection. The qualified packaged virus vector was transfected into CD44+/CD24− cells. The brief steps were as follows: the diluted packaged virus was mixed with the prepared LipoFiterTM solution (Hanbio Biotechnology, Shanghai, China). After incubation at room temperature for 20 minutes, all LipoFiterTM DNA mixture was added to the 12 well plate of cells growing to the logarithmic phase. After shaking, the 12 well plate was transferred to a 37 ℃ and 5% CO2 incubator for continuous culture of cells for 6 hours. The culture medium containing LipoFiterTM DNA was sucked away with a pipette gun. Then, 1 mL of corresponding stem cell culture medium was added to each well and placed in a 37 ℃ and 5% CO2 incubator for continuous culture for 24–40 hours. During and after the culture, the fluorescence in the cells was observed. The transfected cells were screened by puromycin to obtain positively transfected cells, which continued to be cultured for subsequent experiments.
Construction of CD44+/CD24− cell lines with overexpression of CHOP (oeCHOP) and overexpression of PERK (oePERK)
The mRNA sequences of CHOP (NM_001195055) and PERK (NM_004836.7) were obtained from GeneBank, the corresponding open reading frame (ORF) sequences were obtained, and the primers were designed. The primer sequences were CHOP (48800-1)-P1 AGGTCGACTCTAGGATCAGCCGCCACCATGGAGCTTGTTCCAGCCACTCCCC, CHOP (48800-1)-P2 TCCTTGTAGTCCATACCCTTGGTGCAGATTCACATTC; PERK (45005-J1)-P1 AACCGTCAGATCGCACCGGCGCCACCATGGAGCGCGCCATCAGCCCGGGGCTGCTGG, PERK (45005-J1)-P2 TCCTTGTAGTCCATGAATTCATTGCTTGGCAAAGGGCTATGGGAGTTG, the target gene was obtained in the complementary DNA (cDNA) library by qPCR. The obtained target fragment was exchanged into linearized GV208 expression vector, and the recombinant product was transformed into DH5α. After culture on the screening plate, the monoclonal antibodies on the plate were selected for PCR identification, sequencing of the positive clones, and analysis of the results. After obtaining the correct clone, the culture was expanded and extracted, and high-purity plasmids were obtained. Then, the overexpression CD44+/CD24− cell lines of CHOP and PERK were established by lentivirus packaging and cell transfection during the construction of a CHOP gene low expression EC9706 cell line.
Flow cytometry assay to detect cell apoptosis
Each group of cells was treated with 0.25% trypsin to create a single-cell suspension. The cells were then washed with PBS through centrifugation at 1,500 rpm and resuspended in a binding buffer. Next, 5 µL of FITC-labeled annexin V and 10 µL of PI were added to a 200 µL cell suspension containing 5×105 cells/mL. The mixture was incubated at room temperature for 20 minutes, and cell apoptosis was measured using flow cytometry.
Nuclear staining assay
Hoechst 33258 was employed for the purpose of staining the cell nuclei in all groups, with subsequent observation and analysis of nuclear morphology using a fluorescence microscope that operated at an excitation wavelength of 350 nm.
JC-1 assay
The cells in each group were rinsed two times using PBS solution. Then, 1 mL of JC-1 solution, which was prepared using JC-1 staining buffer at a concentration of 10 µg/mL, was added to each well. The cells were placed in a 5% CO2 incubator at a temperature of 37 ℃ for 30 minutes. After that, the cells were washed two times with JC-1 staining buffer and observed under a fluorescence microscope.
Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay
Each group of cells was washed twice using PBS. Then, a solution of 10 mM DCFH-DA in a serum-free medium was added to each well, with a volume of 1 mL. The cells were placed in a 5% CO2 incubator at a temperature of 37 ℃ for 30 minutes. Afterward, the cells were washed twice with PBS and examined under a fluorescence microscope.
Measurement of Cyt C release from mitochondria
The Cyt C releasing apoptosis assay kit was used to separate the mitochondria and the cytosol. To begin, cells were suspended in cytosol extraction buffer and then left to incubate on ice for 10 minutes. Afterwards, the cells were homogenized using a Dounce homogenizer (KIMBLE) and centrifuged at 800 g for 10 minutes. The supernatant obtained was re-centrifuged at 10,000 g for 30 minutes. The resulting supernatant was processed as the cytosolic fraction, while the pellet was processed as the mitochondrial fraction for western blotting (WB) analysis.
RT-PCR assay
Total RNA was extracted from tumor tissues according to TRIeasyTM total RNA extraction reagent kit instruction, and the quality of extracted RNA was confirmed using NanoDrop 1000. Then, cDNA was synthesized using HyperScript III RT SuperMix for qPCR with gDNA Remover Kit. The expression of mRNA was measured by RT-PCR with S6 Universal SYBR qPCR Mix. A two-step process was conducted as follows for RT-PCR: step 1: 1 cycle of 95 ℃ for 30 s; step 2: 40 cycles of 95 ℃ for 10 s, and then 60 ℃ for 30 s. According to the PCR standard amplification curve, the original cycle threshold (Ct) values of the target genes were obtained, and the 2−ΔΔCt method was used for semiquantitative analysis. Table 1 shows the detailed primer sequence of each gene.
Table 1
| Gene name | 5'→3' | Length (bp) |
|---|---|---|
| Caspase 3 | 126 | |
| F | TGCTATTGTGAGGCGGTTGT | |
| R | TCCAGAGTCCATTGATTCGCT | |
| Caspase 6 | 155 | |
| F | GCACCCGGCAGTGTCAA | |
| R | GGCTGTGACACTTGTCTCCTT | |
| Caspase 7 | 168 | |
| F | GGGCCCATCAATGACACAGA | |
| R | GTCTTTTCCGTGCTCCTCCA | |
| Caspase 8 | 118 | |
| F | GAGCCTGAGAGAGCGATGTC | |
| R | AGGCTGAGGCATCTGTTTCC |
F, forward; R, reverse.
WB assay
Cells in each group were washed with PBS, RIPA buffer was added, and the cells were incubated on ice for cell lysis. After the cells were lysed, they were collected into new centrifuge tubes and lysis was continued for 30 minutes. Afterward, the cells were centrifuged at 13,000 rpm, and the supernatants were collected. The protein concentrations were determined. The proteins were added with 4× loading buffer and boiled for 5 minutes until completely denatured. The proteins were then stored at −20 ℃ until use. The gel was prepared by combining 10% separation gel and 5% stacking gel, and the loading volume of protein was determined according to the protein concentration. The proteins were subjected to electrophoresis, membrane transfer, blocking, and incubation with primary antibodies overnight. They were then washed with PBS, incubated with secondary antibodies (at a dilution of 1:2,000) for 1.5 hours at room temperature, washed with PBS again, and visualized with enhanced chemiluminescence (ECL) reagents. X-ray films were developed and photographed. The Image-Pro Plus software (Media Cybernetics, Rockville, MD, USA) was used to analyze the optical density.
Immunofluorescence assay
Cells were directly washed with PBS in 6-well plates. Then, 4% paraformaldehyde was added for 15 minutes for fixation at room temperature, followed by washing with PBS. Afterward, 0.25% Triton was added for 15 minutes for membrane permeabilization, followed incubation with 5% BSA blocking solution prepared in 0.25% Triton for 30 minutes for blocking. After incubation, the blocking solution was aspirated, and the primary antibody prepared in the blocking solution was added. The relative antibodies were diluted in blocking buffer according to the instructions. After incubation overnight at 4 ℃, the cells were washed in PBS, after which DyLight 488-labelled secondary antibodies diluted to 1:2,000 were added. The cells were incubated for 1 hour at room temperature, washed with PBS, and photographed under a fluorescence microscope. The Image-Pro Plus software was used to analyse the optical density.
Statistical analysis
The statistical software SPSS 22.0 (IBM Corp., Armonk, NY, USA) was used to analyze the measurement data. The measurement data were expressed as mean ± standard deviation (SD). If the data conformed to a normal distribution, one-way analysis of variance (ANOVA) was used for comparison among multiple groups, and least significant difference (LSD) was used for pairwise comparison. If the distribution was not normal, the median and interquartile range was used. P<0.05 was considered statistically significant.
Results
Screening and identification of esophageal CSCs
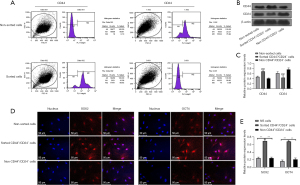
Cells with the CD44+CD24− phenotype in multiple tumors have been proven to serve as tumor stem cell. In this study, magnetic bead sorting technology was used to isolate CD44+CD24− cells from EC. The results demonstrated that CD44+CD24− cells were significantly enriched by flow sorting technology (Figure 1A), and the enrichment efficiency on CD44+CD24− cells was confirmed by detecting the expression of CD44 and CD24 protein in cells (Figure 1B,1C). Further detection of stem cell markers SOX2 and OCT4 in cells showed that the expressions of SOX2 and OCT4 in CD44+CD24− cells were significantly higher than those in non-sorted cells and non-CD44+CD24− cells (P<0.05, Figure 1D,1E), suggesting that CD44+CD24− cells are a group of cells with stem cell characteristics.
Esophageal CSCs have resistance to hypoxia-induced apoptosis
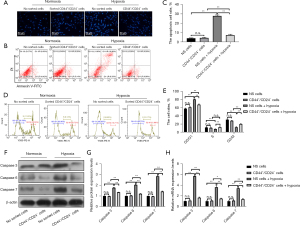
In order to explore the resistance of esophageal CSCs to hypoxia, this study examined the nuclear morphology, cycle distribution, apoptosis ratio, and mitochondrial membrane potential of unsorted EC cells and sorted CD44+/CD24− cells in both normoxic and hypoxic environments. The results showed that under normal culture conditions, compared with CD44+/CD24− cells, the nuclear morphology, cycle distribution, apoptosis ratio, and mitochondrial membrane potential of EC9706 cells did not change significantly. Compared with the unsorted cells cultured in normoxic culture, the nucleus of unsorted cells in hypoxic culture showed obvious shrinkage and lysis, and the cycle was significantly blocked in G0/G1 phase, the proportion of cell apoptosis was increased considerably, and the mitochondrial membrane potential was significantly decreased (all P<0.05). Compared with normal cultured CD44+/CD24− cells, the nuclear morphology, cycle distribution, apoptosis ratio, and mitochondrial membrane potential of CD44+/CD24− cells treated with hypoxia did not change significantly (P>0.05), The above results suggest that CD44+/CD24− cells in EC have stronger resistance and adaptability to hypoxic conditions compared to unsorted EC9706 cells (Figure 2).
Esophageal CSCs can reduce hypoxia-induced apoptosis by inhibiting the mitochondrial apoptosis pathway and ERS-mediated apoptosis pathway
Research has found that there are three pathways of apoptosis in cells: the mitochondrial pathway, the Fas receptor-mediated pathway, and the ERS-mediated pathway (29). To elucidate which apoptosis pathway esophageal CSCs use to resist hypoxia-induced cell apoptosis, this study examined the above three apoptosis pathways in esophageal CSCs in both hypoxic and normoxic environments.
Effects of hypoxia on mitochondrial apoptosis pathways in EC cells
Compared with CD44+/CD24− cells, the expression levels of Bax, Apaf, caspase 9, and Bcl-2 proteins in EC9706 cells were not significantly changed (P>0.05). The expression levels of Bax, Apaf, and caspase 9 were significantly increased (P<0.05) and Bcl-2 was significantly decreased (P<0.05) in unsorted cells after hypoxia treatment compared with normal cultured unsorted cells. Compared with CD44+/CD24− cells after normal culture, The protein expression levels of Bax, Apaf, caspase 9, and Bcl-2 in CD44+/CD24− cells treated with hypoxia did not change significantly (P>0.05) (Figure 3A,3B).
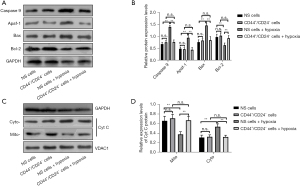
In addition, the protein content of Cyt C in mitochondria and cytoplasm of EC9706 cells was further detected, and it was found that the content of Cyt C in mitochondria and cytoplasm of EC9706 cells was significantly decreased (P<0.05), and the protein content of Cyt C in the cytoplasm was significantly increased (P<0.05) after hypoxia induction. However, there was no significant change of Cyt C in mitochondria and cytoplasm of CD44+/CD24− cells after hypoxia induction (P>0.05) (Figure 3C,3D).
Effect of hypoxia on Fas/Fas ligand (FasL) apoptotic pathway in EC cells
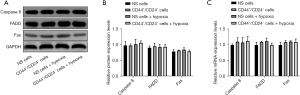
Compared with CD44+/CD24− cells, the contents of FAS, caspase 8, and FADD proteins in EC9706 cells did not change significantly after hypoxia treatment (P>0.05). At the mRNA level, the variation trend of the above factors in each group was consistent with the protein level (Figure 4).
Effects of hypoxia on ERS-mediated apoptosis pathway in EC cells
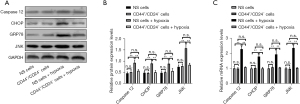
Compared with CD44+/CD24− cells, the expressions of GRP78, caspase 12, CHOP, and JNK proteins in EC9706 cells had no significant changes (P>0.05). The protein expressions of GRP78, caspase 12, CHOP, and JNK in unsorted cells after hypoxia treatment were significantly increased compared with those in normal cultured unsorted cells (P<0.05), and compared with CD44+/CD24− cells after normal culture, the protein expressions of GRP78, caspase 12, CHOP, and JNK in CD44+/CD24− cells were not significantly changed after hypoxia treatment (P>0.05). At the mRNA level, the variation trend of the above factors in each group was consistent with the protein level (Figure 5). The above results indicate that CD44+/CD24− cells can inhibit the activation of the hypoxia-induced ERS pathway and the activation of the hypoxia-induced mitochondrial apoptosis pathway. This endows CD44+/CD24− cells with the ability to resist hypoxic conditions by suppressing the activation of the ERS pathway and mitochondrial apoptosis pathway in a low oxygen environment.
Esophageal CSCs resist hypoxia-induced apoptosis by inhibiting the activation of CHOP on mitochondrial apoptosis pathway
Currently, research has found that the CHOP protein is a key factor in inducing cellular apoptosis through the ERS pathway. The CHOP protein acts as a bridge between the ERS apoptosis pathway and the mitochondrial apoptosis pathway. Therefore, this study hypothesizes that hypoxia-induced apoptosis in EC9706 cells is achieved through the activation of the ERS pathway, followed by activation of the mitochondrial apoptosis pathway through the ERS pathway. Conversely, one of the mechanisms by which CD44+CD24− tumor stem cells resist hypoxia-induced apoptosis may be achieved through the inhibition of the activation of the ERS pathway. To confirm this hypothesis, this study silenced the CHOP gene in EC9706 cells and overexpressed it in CD44+CD24− cells, and then tested the anti-apoptotic ability of the unsorted EC cells and sorted CD44+CD24− cells under normoxia and hypoxia conditions.
Construction of low expression CHOP EC9706 cell lines and overexpression CHOP CD44+/CD24− cell lines
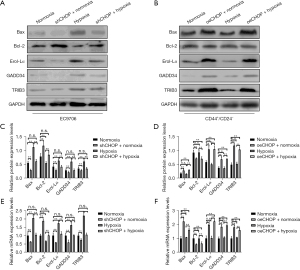
WB and PCR assays results showed that the low-expression CHOP EC9706 cell and the overexpression CHOP CD44+/CD24− cell lines had been successfully constructed (Figure 6).

Effects of silencing or oeCHOP on cell cycle and apoptosis induced by hypoxia
Apoptosis
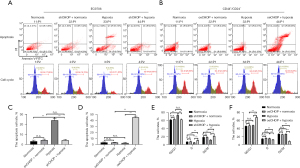
Compared with EC9706 cells cultured under normal conditions, apoptosis of EC9706 cells increased significantly after hypoxia induction, and apoptosis of cells cultured under normoxic conditions shRNA-CHOP group and cells cultured under hypoxia induction shRNA-CHOP group had no significant change; compared with CD44+/CD24− cells cultured under normal conditions, the apoptosis of CD44+/CD24− cells cultured under hypoxia and oeCHOP under normoxia showed no significant change, but the apoptosis of oeCHOP under hypoxia increased significantly (Figure 7A-7D).
Cycle arrest
Compared with EC9706 cells under normal culture conditions, EC9706 cells were significantly arrested in G0/G1 phase after hypoxia induction, and the cell cycle of the shRNA-CHOP group under normoxic conditions and shRNA-CHOP group after hypoxia induction had no significant change. Compared with CD44+/CD24− cells cultured under normal conditions, there was no significant change in the cycle of CD44+/CD24− cells after hypoxia induction and in the oeCHOP group under normoxic conditions, and the cell cycle of the oeCHOP group under hypoxic conditions was significantly inhibited to the G0/G1 phase (Figure 7A,7B,7E,7F).
Effects of silencing or oeCHOP on mitochondrial membrane potential induced by hypoxia
Compared with EC9706 cells cultured under normal conditions, the mitochondrial membrane potential of EC9706 cells decreased significantly after hypoxia induction, and there was no significant change in shRNA-CHOP cells under normoxia and shRNA-CHOP cells after hypoxia induction. Compared with CD44+/CD24− cells cultured under normal conditions, the mitochondrial membrane potential of CD44+/CD24− cells cultured under hypoxia and oeCHOP under normoxia did not change significantly. Still, the mitochondrial membrane potential of cells cultured under hypoxia oeCHOP increased significantly (Figure 8).
Effects of silencing or oeCHOP on the expression of apoptosis-related proteins Bax, Bcl-2, GADD34, Ero1-Lα, and TRIB3 in cells induced by hypoxia
Compared with EC9706 cells under normal culture conditions, the protein expression of Bax, GADD34, Ero1-Lα, and TRIB3 in the shRNA-CHOP group was significantly decreased, and the protein expression of Bcl-2 was significantly increased in shRNA-CHOP group under normoxic conditions. The expression of Bax, GADD34, Ero1-Lα, and TRIB3 in EC9706 cells increased dramatically after hypoxia induction, whereas the expression of Bcl-2 was decreased significantly after hypoxia induction. The expression of Bax, Bcl-2, GADD34, Ero1-Lα, and TRIB3 was not considerably changed in the shRNA-CHOP group after hypoxia induction. Compared with CD44+/CD24− cells under normal culture conditions, the protein expressions of Bax, GADD34, Ero1-Lα, and TRIB3 were significantly increased, and Bcl-2 was significantly decreased in the oeCHOP group under normoxic conditions. The protein expressions of Bax, Bcl-2, GADD34, Ero1-Lα, and TRIB3 in CD44+/CD24− cells were not significantly changed after hypoxia induction. Still, the protein expressions of Bax, GADD34, Ero1-Lα, and TRIB3 were increased considerably in the oeCHOP group under hypoxia. Bcl-2 protein expression was significantly decreased. At the mRNA level, the variation trend of the above factors in each group was consistent with the protein level (Figure 9).
Effects of mitochondrial membrane permeability inhibitors on apoptosis, mitochondrial membrane potential, and mitochondrial apoptosis related proteins Bax, Bcl-2, Apaf, Cyt C, and caspase 9 of hypoxia-induced CD44+/CD24− cells with oeCHOP
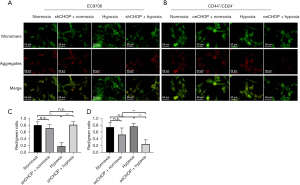
Compared with the sorting group, the apoptosis ratio of the oeCHOP + hypoxia group was significantly increased, the mitochondrial membrane potential was significantly decreased, the protein expressions of Bax, Apaf, Cyt C, and caspase 9 were significantly increased, and the expression of Bcl-2 was significantly decreased. Compared with the oeCHOP + hypoxia group, the apoptosis ratio of oeCHOP + hypoxia + mitochondrial membrane permeability inhibitor group was significantly decreased, the mitochondrial membrane potential was significantly increased, the protein expressions of Bax, Apaf, Cyt C, and caspase 9 were reduced considerably, and the expression of Bcl-2 was significantly increased (Figure 10).
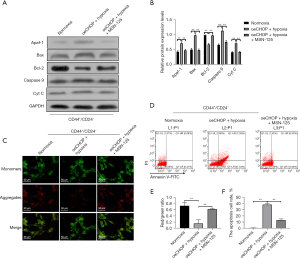
The above results further suggest that the molecular mechanism of CD44+CD24− cells resisting hypoxia is associated with their ability to suppress CHOP-induced mitochondrial apoptosis.
Effect of hypoxia on the expression of GRP78, PERK, eIF2α, and ATF4 proteins in esophageal CSCs
Research has found that when cells undergo ERS, unfolded proteins bind to GRP78, leading to the activation of the PERK-eIF2α-ATF4 signaling pathway. As a result, the expression of CHOP, a target gene of ATF4, increases. oeCHOP then induces cell apoptosis. Therefore, this study speculates that hypoxia induces apoptosis in EC cells through the activation of the GRP78-PERK-eIF2α-ATF4-CHOP pathway, while esophageal CSCs can resist apoptosis by inhibiting the high expression of CHOP induced by hypoxia. Based on this, this study further explores the molecular mechanisms of CHOP-mediated resistance to hypoxia in CD44+CD24− cells.
Compared with CD44+/CD24− cells, there was no significant change in the mRNA expression of GRP78, PERK, eIF2α, and ATF4 in EC9706 cells under normoxic culture (P>0.05). The mRNA expressions of GRP78, perk, eIF2α, and ATF4 in non-sorted cells after hypoxia treatment were significantly higher than those in normal cultured non-sorted control cells (P<0.05). Compared with normal cultured CD44+/CD24− cells, the mRNA expressions of GRP78, PERK, eIF2α, and ATF4 in CD44+/CD24− cells after hypoxia treatment did not change significantly (P>0.05) (Figure 11).
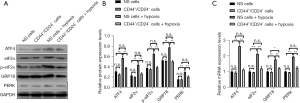
At the protein level, compared with CD44+/CD24− cells, EC9706 cells had higher levels of GRP78, PERK, eIF2α, and p-eIF2α. There was no significant change in the expression of ATF4 protein (P>0.05). Compared with the normal cultured unselected control cells, GRP78, PERK, p-eIF2α, and eIF2α in the unselected cells after hypoxia treatment the expression of ATF4 protein increased significantly (P<0.05). Compared with normal cultured CD44+/CD24− cells, GRP78, PERK, p-eIF2α, and eIF2α in CD44+/CD24− cells treated with hypoxia, the expression of ATF4 protein did not change significantly (P>0.05) (Figure 11).
Esophageal CSCs inhibit hypoxia-induced apoptosis by inhibiting the activation of the GRP78-PERK-eIF2α-ATF4 signaling pathway
To further confirm the role of the GRP78-PERK-eIF2α-ATF4-CHOP signaling pathway in the inactivation of esophageal CSCs under hypoxic conditions, this study further investigated the anti-apoptotic capability of unsorted EC cells and sorted CD44+/CD24− cells under normoxic and hypoxic conditions by silencing and overexpressing the key molecule PERK in GRP78-PERK-eIF2α-ATF4-CHOP pathway.
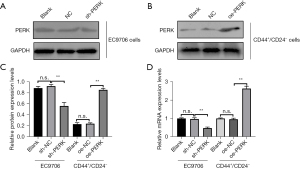
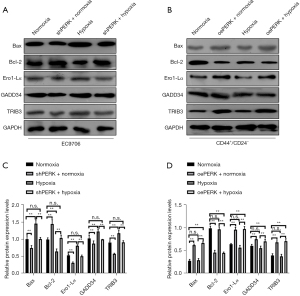
Construction of low-expression PERK EC9706 cell lines and overexpression PERK CD44+/CD24− cell lines
The results of WB and PCR assays showed that the low-expression PERK EC9706 cell and the overexpression PERK CD44+/CD24− cell lines had been successfully constructed (Figure 12).
Effects of silencing or oePERK gene on cell cycle, apoptosis, and mitochondrial membrane potential of cells induced by hypoxia
Apoptosis: compared with EC9706 cells under normal culture conditions, EC9706 cell apoptosis increased significantly after hypoxia induction (P<0.05), and there was no significant change in shRNA PERK group cells under normoxia and shRNA PERK group cells after hypoxia induction. Compared with CD44+/CD24− cells under normal culture conditions, there was no significant change in CD44+/CD24− cell apoptosis induced by hypoxia and oePERK group under normoxia, but the apoptosis of oePERK group under hypoxia increased significantly (P<0.05) (Figure 13A-13D).
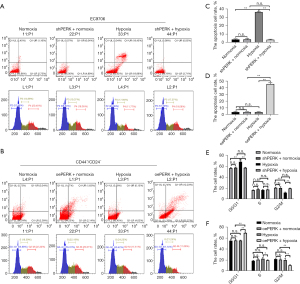
Cycle arrest: compared with EC9706 cells under normal culture conditions, EC9706 cells were significantly blocked in G0/G1 phase after hypoxia induction (P<0.05). There was no significant change in the cell cycle of shRNA PERK EC9706 group under normoxia and shRNA PERK EC9706 group after hypoxia induction. Compared with CD44+/CD24− cells under normal culture conditions, the apoptosis of CD44+/CD24− cells induced by hypoxia and the cell cycle of OE perk CD44+/CD24− group under normoxia had no significant change, and the cell cycle of oePERK CD44+/CD24− group under hypoxia was significantly inhibited to G0/G1 phase (P<0.05) (Figure 13A,13B,13E,13F).
The changing trend of mitochondrial membrane potential was consistent with the above (Figure 14).
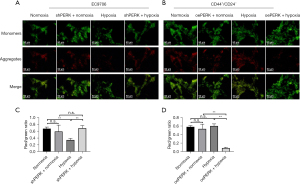
Effects of silencing or oePERK on the expression of apoptosis-related proteins Bax, Bcl-2, GADD34, Ero1-Lα, and TRIB3 in cells induced by hypoxia
Compared with EC9706 cells under normal culture conditions, the expression of TRIB3, Bax, GADD34, and Ero1-Lα protein in shRNA PERK group cells under normoxic conditions decreased significantly (P<0.05), and the expression of Bcl-2 protein increased significantly (P<0.05). The expression of TRIB3, Bax, GADD34, and Ero1-Lα protein in EC9706 cells induced by hypoxia increased significantly (P<0.05), and Bcl-2 protein decreased significantly (P<0.05). The expression of TRIB3 protein Bax, Bcl-2, GADD34, and Ero1-Lα in the shRNA PERK EC9706 group induced by hypoxia had no significant change. Compared with CD44+/CD24− cells cultured under normal conditions, the expression of BAX, GADD34, Ero1-Lα, and TRIB3 proteins in the oePERK group cells under normoxic conditions significantly increased (P<0.05), while Bcl-2 significantly decreased (P<0.05). After hypoxia induction, there was no significant change in the expression of BAX, Bcl-2, GADD34, Ero1-Lα, and TRIB3 proteins in CD44+/CD24− cells. Under hypoxic conditions, the expression of BAX, GADD34, Ero1-Lα, and TRIB3 proteins in the oePERK group cells increased significantly (P<0.05), while the expression of Bcl-2 protein decreased significantly (P<0.05) (Figure 15).
The above results further suggested that the inactivation of the GRP78-PERK-eIF2α-ATF4-CHOP signaling pathway in esophageal CSCs under hypoxic conditions may be the main molecular mechanism contributing to their resistance to hypoxia-induced apoptosis.
Discussion
At present, studies have found that the resistance of CSCs to hypoxia-induced apoptosis is related to their own hypoxic microenvironment and metabolic state (30). CSCs exist in a hypoxic microenvironment caused by the disharmony between angiogenesis rate and tumor growth rate (31,32), which allows them to obtain the ability to resist apoptosis by changing the glucose metabolic pathway and enhancing the glycolysis process of cells (33,34). However, relying solely on changing metabolic pathways to maintain low levels of proliferation and differentiation and other positive factors that promote cell survival does not seem to completely explain how tumor stem cells evade the accumulation of errors in protein synthesis or folding caused by ERS and mitochondrial membrane damage-induced apoptosis under long-term hypoxic conditions. Therefore, there may be an intrinsic signaling system in esophageal CSCs that ensures their survival under prolonged hypoxia. To explore the molecular mechanism of anti-hypoxia-induced apoptosis of esophageal CSCs, the CD44+/CD24− subpopulation were isolated from EC9706 cells. Through verification, it was found that compared with non-sorted EC9706 cells, the expression level of stem cell marker proteins OCT4 and SOX2 increased significantly in CD44+/CD24− cells, suggesting that CD44+/CD24− cells have CSC characteristics. After hypoxic treatment of unsorted EC9706 cells and CD44+/CD24− cells, it was found that compared with EC9706 cells in the normoxic conditions, EC9706 cells in the hypoxic conditions showed obvious nuclear pyknosis and karyolysis, the significant increase in proportion of apoptosis, cell cycle arrest in G0/G1 phase, and the significant loss in the mitochondrial membrane potential. However, the nuclear morphology, apoptosis ratio, cycle distribution, and mitochondrial membrane potential of CD44+/CD24− cells under hypoxia condition did not change significantly. At the same time, hypoxia can induce a significant increase in the protein and mRNA levels of caspase 3, caspase 6, and caspase 7 in unsorted EC9706 cells, but the level of caspase 3, caspase 6, and caspase 7 protein and mRNA in CD44+/CD24− cells is not affected by hypoxia, remaining at a low level. As key executor proteins of apoptosis, the activated caspase 3, caspase 6, and caspase 7 hydrolyzes the target protein, resulting in the cell entering into the process of apoptosis (35,36). These results suggest that CD44+/CD24− CSCs in EC cell line EC9706 have strong resistance to hypoxia and can resist hypoxia-induced apoptosis.
Apoptosis pathways include the mitochondrial apoptosis pathway, Fas/FasL pathway, and ERS pathway (37,38), which are closely related to the occurrence and development of cancer and can initiate apoptosis (39). At present, there is no evidence to confirm which apoptosis pathway esophageal CSCs regulate to inhibit hypoxia-induced apoptosis. This study found that hypoxia did not affect the Fas/FasL apoptosis pathway in EC9706 cells and CD44+/CD24− cells. Still, hypoxia could significantly increase the expression of Bax, Apaf, and caspase 9 protein, significantly reduce Bcl-2, and activate the mitochondrial apoptosis pathway of EC9706 cells. At the same time, hypoxia had no significant effect on the mitochondrial apoptosis of sorted CD44+/CD24− cells. This suggests that hypoxia may induce apoptosis by activating the mitochondrial apoptosis pathway in EC9706 cells rather than Fas/FasL apoptosis pathway. CD44+/CD24− cells can resist hypoxia-induced apoptosis by inhibiting the activation of the mitochondrial apoptosis pathway. Continuous hypoxia will also activate the mitochondrial pathway through ERS pathway and induce apoptosis (40-44). This study further found that hypoxia can induce the expression of ERS pathway marker proteins GRP78, caspase 12, CHOP, and JNK in EC9706 cells, but has no effect on the expression of GRP78, caspase 12, CHOP, and JNK in CD44+/CD24− cells. As the marker proteins of ERS pathway, the above proteins play an essential role in the activation of ERS pathway (23,25,45). Based on the findings of this study, CD44+/CD24− cells can inhibit the activation of hypoxia-induced ERS pathway and hypoxia-induced mitochondrial apoptosis pathway. This study preliminarily confirmed that one of the mechanisms of CD44+/CD24− cells resisting hypoxia-induced apoptosis might be realized by inhibiting the activation of ERS pathway and then inhibiting the mitochondrial apoptosis pathway.
Research has found that CHOP protein is the key factor of ERS pathway-induced apoptosis (36,46,47). ERS induces the increase of CHOP expression, which can inhibit the transcription of Bcl-2, resulting in the relative excess of Bax (23). The increased free Bax is transported to mitochondria and induce mitochondrial pathway apoptosis (26). This study found that silencing CHOP in EC9706 cells can significantly reduce hypoxia-induced apoptosis of EC9706 cells, relieve the cycle arrest caused by hypoxia, improve the reduced mitochondrial membrane potential, and increase the ability of EC9706 cells to resist hypoxia-induced apoptosis. oeCHOP in CD44+/CD24− cells can significantly enhance hypoxia-induced apoptosis of CD44+/CD24− cells, increase cycle arrest, and decrease mitochondrial membrane potential, which weakens the resistance of CD44+/CD24− cells to hypoxia-induced apoptosis. This further confirmed that the molecular mechanism of CD44+/CD24− cells against hypoxia was related to the inhibition of activation of ERS pathway to mitochondrial apoptosis.
At present, it has been found that overexpressed CHOP can accelerate p-eIF2α by promoting the expression of GADD34 dephosphorylation, inhibit the transcription of Bcl-2, upregulate Ero1-Lα and TRIB3 expression, then induces apoptosis (25-27,48-50). Therefore, this study detected the downstream apoptosis-related factors regulated by CHOP under normoxia and hypoxia. The results showed that silencing CHOP could significantly inhibit an increase in the protein expression of Bax, GADD34, Ero1-Lα, and TRIB3, and a decrease in the level of Bcl-2 in hypoxia-induced EC9706 cells. oeCHOP could significantly promote the protein expression of Bax, GADD34, Ero1-Lα, and TRIB3, and inhibit the expression of Bcl-2 in CD44+/CD24− cells under hypoxic conditions. The above further confirmed that hypoxia could activate the ERS apoptosis pathway in EC9706 cells, then activate the mitochondrial apoptosis pathway and induce apoptosis. Still, hypoxia does not activate the ERS apoptosis pathway in CD44+/CD24− tumor stem cells selected from EC9706 cells. CD44+/CD24− tumor stem cells can resist hypoxia-induced apoptosis by inhibiting the activation of ERS by hypoxia. Whether CD44+/CD24− CSCs inhibit mitochondrial apoptosis pathway through the ERS pathway and resist hypoxia-induced apoptosis was further studied. The results showed that Bax oligomer inhibitor MSN-125 could reverse the increase of apoptosis induced by hypoxia and inhibit the activation of hypoxia-induced mitochondrial apoptosis pathway in CD44+/CD24− cells overexpressing CHOP.
Previous studies have found that CHOP, as one of the main factors in ERS mediated apoptosis pathway, activates the receptor protein PERK on the endoplasmic reticulum membrane when the cells experience continuous or too strong ERS and too many unfolded proteins bind to GRP78 (51,52). The activated PERK phosphorylates eIF2α and p-eIF2α to inhibit protein translation and selectively activate the transcription and translation of ATF4 (23), and CHOP, as the target gene of ATF4, increases. oeCHOP further induces apoptosis (53). Therefore, this study was based on the GRP78-PERK-eIF2α-ATF4-CHOP signaling pathway to further explore the molecular mechanism of hypoxia-induced apoptosis of EC cells by activating CHOP-mediated apoptosis, whereas esophageal CSCs could resist apoptosis by inhibiting the high expression of CHOP induced by hypoxia. This study first found that hypoxia could induce perk, GRP78, and eIF2α in EC9706 cells and the protein and mRNA expression of ATF4 increased and promoted eIF2. However, it was shown to have no effect on PERK, GRP78, and eIF2α in CD44+/CD24− cells and ATF4 protein and mRNA and eIF2α. This preliminarily suggests that esophageal CSCs having no effect on the inactivation of the GRP78-PERK-EIF2α-ATF4-CHOP signaling pathway under hypoxia may be the primary molecular mechanism of its anti-hypoxia induced apoptosis. In order to further confirm the above conclusions, this study further silenced perk in EC9706 cells, overexpressed perk in CD44+/CD24− cells, and then intervened with hypoxia. The results showed that silencing PERK in EC9706 cells could significantly reduce hypoxia-induced apoptosis of EC9706 cells, relieve the cycle arrest caused by hypoxia, and improve the reduced mitochondrial membrane potential; it increased the ability of EC9706 cells to resist hypoxia-induced apoptosis and oePERK in CD44+/CD24− cells could significantly increase hypoxia-induced CD44+/CD24− cell apoptosis, increase cycle arrest, reduce mitochondrial membrane potential, and then weaken the resistance of CD44+/CD24− cells to hypoxia-induced apoptosis. Further study on the detection of apoptosis-related factors Bcl-2, Bax, GADD34, Ero1-Lα, and TRIB3 confirmed that PERK silencing enhanced the ability of EC9706 cells to resist hypoxia-induced apoptosis, and that PERK overexpression decreased the ability of CD44+/CD24− cells to resist hypoxia-induced apoptosis. At the same time, the ability of GRP78-PERK-eIF2α-ATF4 transcription factors in the middle and lower reaches of ATF4-CHOP signal pathway to regulate chop expression was detected by a double luciferase reporting experiment. It was found that perk silencing significantly weakened the binding ability of ATF4 to chop promoter of EC9706 cells under hypoxia-induced conditions, and PERK overexpression greatly enhanced the binding capacity of ATF4 to the CHOP promoter of CD44+/CD24− cells under hypoxia-induced conditions. The above experiments confirmed that esophageal CSCs could inhibit hypoxia. The activation of GRP78-PERK-eIF2α-ATF4-CHOP signaling pathway can obtain the ability to resist hypoxia-induced apoptosis.
Conclusions
In conclusion, CD44+/CD24− CSCs in EC cell line EC9706 can resist hypoxia-induced apoptosis. The molecular mechanism of this resistance is that CD44+/CD24− tumor stem cells can inhibit CHOP-mediated ERS apoptosis pathway and further inhibit the activation of the mitochondrial apoptosis pathway.
Acknowledgments
We thank Dr. Beatrice Aramini (Department of Medical and Surgical Sciences-DIMEC of the Alma Mater Studiorum-University of Bologna, Bologna, Italy) for the critical comments and valuable advice on this study.
Funding: This work was supported by the Natural Science Foundation of Gansu Province (No. 21JR1RA118), Youth Science and Technology Fund Program of Gansu Province (No. 21JR1RA107), and the Foundation of The First Hospital of Lanzhou University, China (Nos. ldyyyn2021-8 and ldyyyn2021-59).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-462/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-462/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-462/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-462/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Short MW, Burgers KG, Fry VT. Esophageal Cancer. Am Fam Physician 2017;95:22-8. [PubMed]
- Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg 2018;41:210-5. [Crossref] [PubMed]
- Gai L, Huang Y, Zhao L, et al. Long non-coding RNA HAGLROS regulates the proliferation, migration, and apoptosis of esophageal cancer cells via the HAGLROS-miR-206-NOTCH3 axis. J Gastrointest Oncol 2021;12:2093-108. [Crossref] [PubMed]
- Lohan-Codeço M, Barambo-Wagner ML, Nasciutti LE, et al. Molecular mechanisms associated with chemoresistance in esophageal cancer. Cell Mol Life Sci 2022;79:116. [Crossref] [PubMed]
- Liu K, Zhao T, Wang J, et al. Etiology, cancer stem cells and potential diagnostic biomarkers for esophageal cancer. Cancer Lett 2019;458:21-8. [Crossref] [PubMed]
- Izadpanah MH, Forghanifard MM. TWIST1 Plays Role in Expression of Stemness State Markers in ESCC. Genes (Basel) 2022;13:2369. [Crossref] [PubMed]
- Najafi M, Mortezaee K, Majidpoor J. Cancer stem cell (CSC) resistance drivers. Life Sci 2019;234:116781. [Crossref] [PubMed]
- Zhou C, Fan N, Liu F, et al. Linking Cancer Stem Cell Plasticity to Therapeutic Resistance-Mechanism and Novel Therapeutic Strategies in Esophageal Cancer. Cells 2020;9:1481. [Crossref] [PubMed]
- Li MY, Fan LN, Han DH, et al. Ribosomal S6 protein kinase 4 promotes radioresistance in esophageal squamous cell carcinoma. J Clin Invest 2020;130:4301-19. [Crossref] [PubMed]
- Trevellin E, Pirozzolo G, Fassan M, et al. Prognostic value of stem cell markers in esophageal and esophagogastric junction cancer: a meta-analysis. J Cancer 2020;11:4240-9. [Crossref] [PubMed]
- Dong Y, Wu Y, Zhao GL, et al. Inhibition of autophagy by 3-MA promotes hypoxia-induced apoptosis in human colorectal cancer cells. Eur Rev Med Pharmacol Sci 2019;23:1047-54. [PubMed]
- Jing X, Yang F, Shao C, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer 2019;18:157. [Crossref] [PubMed]
- Li S, Xu HX, Wu CT, et al. Angiogenesis in pancreatic cancer: current research status and clinical implications. Angiogenesis 2019;22:15-36. [Crossref] [PubMed]
- Ramjiawan RR, Griffioen AW, Duda DG. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy? Angiogenesis 2017;20:185-204. [Crossref] [PubMed]
- Liu Z, Xu J, Liu M, et al. Efficacy and safety of angiogenesis inhibitors plus immune checkpoint inhibitors in advanced soft tissue sarcoma: a real-world, single-center study. Sci Rep 2023;13:3385. [Crossref] [PubMed]
- Chen J, Chen S, Zhuo L, et al. Regulation of cancer stem cell properties, angiogenesis, and vasculogenic mimicry by miR-450a-5p/SOX2 axis in colorectal cancer. Cell Death Dis 2020;11:173. [Crossref] [PubMed]
- Huang T, Song X, Xu D, et al. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics 2020;10:8721-43. [Crossref] [PubMed]
- Li L, Li JC, Yang H, et al. Expansion of cancer stem cell pool initiates lung cancer recurrence before angiogenesis. Proc Natl Acad Sci U S A 2018;115:E8948-57. [Crossref] [PubMed]
- Huang Z, Zhou M, Wang Q, et al. Mechanical and hypoxia stress can cause chondrocytes apoptosis through over-activation of endoplasmic reticulum stress. Arch Oral Biol 2017;84:125-32. [Crossref] [PubMed]
- Zhou Y, Jin Y, Wang Y, et al. Hypoxia activates the unfolded protein response signaling network: An adaptive mechanism for endometriosis. Front Endocrinol (Lausanne) 2022;13:945578. [Crossref] [PubMed]
- Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci 2015;40:141-8. [Crossref] [PubMed]
- Bastida-Ruiz D, Aguilar E, Ditisheim A, et al. Endoplasmic reticulum stress responses in placentation - A true balancing act. Placenta 2017;57:163-9. [Crossref] [PubMed]
- Rozpedek W, Pytel D, Mucha B, et al. The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med 2016;16:533-44. [Crossref] [PubMed]
- DeGracia DJ, Kumar R, Owen CR, et al. Molecular pathways of protein synthesis inhibition during brain reperfusion: implications for neuronal survival or death. J Cereb Blood Flow Metab 2002;22:127-41. [Crossref] [PubMed]
- Lu M, Lawrence DA, Marsters S, et al. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 2014;345:98-101. [Crossref] [PubMed]
- Tamaki T, Kamatsuka K, Sato T, et al. A novel transmembrane protein defines the endoplasmic reticulum stress-induced cell death pathway. Biochem Biophys Res Commun 2017;486:149-55. [Crossref] [PubMed]
- Li Y, Zhu D, Hou L, et al. TRB3 reverses chemotherapy resistance and mediates crosstalk between endoplasmic reticulum stress and AKT signaling pathways in MHCC97H human hepatocellular carcinoma cells. Oncol Lett 2018;15:1343-9. [PubMed]
- Zhao Y, Yu Y, Li H, et al. FAM175B promotes apoptosis by inhibiting ATF4 ubiquitination in esophageal squamous cell carcinoma. Mol Oncol 2019;13:1150-65. [Crossref] [PubMed]
- Gong J, Xie XK. Effect of endoplasmic reticulum stress and autophagy on hepatocyte apoptosis. Journal of Clinical Hepatology 2019;35:2828-32.
- Emami Nejad A, Najafgholian S, Rostami A, et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: a novel approach to developing treatment. Cancer Cell Int 2021;21:62. [Crossref] [PubMed]
- Kim H, Lin Q, Glazer PM, et al. The hypoxic tumor microenvironment in vivo selects the cancer stem cell fate of breast cancer cells. Breast Cancer Res 2018;20:16. [Crossref] [PubMed]
- Höckel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst 2001;93:266-76. [Crossref] [PubMed]
- Sun X, Lv X, Yan Y, et al. Hypoxia-mediated cancer stem cell resistance and targeted therapy. Biomed Pharmacother 2020;130:110623. [Crossref] [PubMed]
- Heddleston JM, Li Z, McLendon RE, et al. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009;8:3274-84. [Crossref] [PubMed]
- Van Opdenbosch N, Lamkanfi M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019;50:1352-64. [Crossref] [PubMed]
- Fan TJ, Han LH, Cong RS, et al. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai) 2005;37:719-27. [Crossref] [PubMed]
- Jeong SY, Seol DW. The role of mitochondria in apoptosis. BMB Rep 2008;41:11-22. [Crossref] [PubMed]
- Waring P, Müllbacher A. Cell death induced by the Fas/Fas ligand pathway and its role in pathology. Immunol Cell Biol 1999;77:312-7. [Crossref] [PubMed]
- Goldar S, Khaniani MS, Derakhshan SM, et al. Molecular mechanisms of apoptosis and roles in cancer development and treatment. Asian Pac J Cancer Prev 2015;16:2129-44. [Crossref] [PubMed]
- Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol 2018;19:109-20. [Crossref] [PubMed]
- Akman M, Belisario DC, Salaroglio IC, et al. Hypoxia, endoplasmic reticulum stress and chemoresistance: dangerous liaisons. J Exp Clin Cancer Res 2021;40:28. [Crossref] [PubMed]
- Koritzinsky M, Levitin F, van den Beucken T, et al. Two phases of disulfide bond formation have differing requirements for oxygen. J Cell Biol 2013;203:615-27. [Crossref] [PubMed]
- Fribley A, Zhang K, Kaufman RJ. Regulation of apoptosis by the unfolded protein response. Methods Mol Biol 2009;559:191-204. [Crossref] [PubMed]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 2011;13:184-90. [Crossref] [PubMed]
- Hu H, Tian M, Ding C, et al. The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Front Immunol 2019;9:3083. [Crossref] [PubMed]
- Lei Y, Wang S, Ren B, et al. CHOP favors endoplasmic reticulum stress-induced apoptosis in hepatocellular carcinoma cells via inhibition of autophagy. PLoS One 2017;12:e0183680. [Crossref] [PubMed]
- Qiu LZ, Yue LX, Ni YH, et al. Emodin-Induced Oxidative Inhibition of Mitochondrial Function Assists BiP/IRE1α/CHOP Signaling-Mediated ER-Related Apoptosis. Oxid Med Cell Longev 2021;2021:8865813. [Crossref] [PubMed]
- Sun J, Wang W, Tian Y, et al. Endoplasmic reticulum stress related reactive oxygen species and its mechanism. China Medical Herald 2018;15:42-4, 54.
- Jin Y, Nguyen TLL, Myung CS, et al. Ginsenoside Rh1 protects human endothelial cells against lipopolysaccharide-induced inflammatory injury through inhibiting TLR2/4-mediated STAT3, NF-κB, and ER stress signaling pathways. Life Sci 2022;309:120973. [Crossref] [PubMed]
- He X, Wu D, Xu Y, et al. Perfluorooctanoic acid promotes pancreatic β cell dysfunction and apoptosis through ER stress and the ATF4/CHOP/TRIB3 pathway. Environ Sci Pollut Res Int 2022;29:84532-45. [Crossref] [PubMed]
- Verfaillie T, Rubio N, Garg AD, et al. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ 2012;19:1880-91. [Crossref] [PubMed]
- Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta 2013;1833:3460-70. [Crossref] [PubMed]
- Li Y, Guo Y, Tang J, et al. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim Biophys Sin (Shanghai) 2014;46:629-40. [Crossref] [PubMed]


