
CircRHOT1 restricts gastric cancer cell ferroptosis by epigenetically regulating GPX4
Highlight box
Key findings
• We found elevated level of circRHOT1 in patients with gastric cancer and identified that circRHOT1 promotes the growth of gastric cancer via recruiting lysine acetyltransferase KAT5 to elevate the expression of ferroptosis suppressor GPX4.
What is known and what is new?
• The GPX4 is a critical regulator of ferroptosis, and circRHOT1 has been identified as potential diagnostic factor for cancers.
• CricRHOT1 epigenetically regulate GPX4 expression to modulate ferroptosis and growth of gastric cancer.
What is the implication, and what should change now?
• CircRHOT1 is a potential diagnostic indicator and a promising therapeutic target for gastric cancer. Cancer treatments that target circRHOT1 should be studied in future.
IntroductionOther Section
Gastric cancer (GC) is one of the most malignant and prevalent types of cancer worldwide (1,2). Patients with GC usually have a poor prognosis and high mortality (3). To date, the mainstream treatments for GC are surgical resection with perioperative chemotherapy for early-stage disease and palliative chemotherapy, with or without immunotherapy or trastuzumab, for advanced stage disease (4). Genetic and epigenetic regulations, such as the transcriptional factors and non-coding RNAs, have been reported to control the growth, death, and metabolism of GC (5,6). Surgical operation is the primary choice for GC patients, and the chemical therapy and radiotherapy are commonly adopted for adjuvant treatment before and after surgery (7). Besides, immunotherapy such as immune checkpoint inhibitors for GC has emerged (8). Despite great efforts and major advances in GC treatments, the overall survival rate and prognosis of GC patients remain poor due to frequent recurrence and tumor metastasis (9). Deciphering the mechanisms and identifying novel biomarkers is crucial for the development of novel and efficient therapies for GC.
Ferroptosis is a novel form of programmed cell death that was only defined in recent years (10). Unlike other traditional regulated cell death progresses, such as apoptosis and necrosis, ferroptosis is characterized by abnormal iron flux, iron-dependent lipid peroxidation, and oxidative cell death (11). The canonical mechanism that induces ferroptosis is the inactivation of the anti-peroxidation mechanisms of the cell membranes, and glutathione peroxidases (GPXs) play critical role in this process (12,13). Glutathione peroxidase-4 (GPX4) is capable of detoxifying hydroperoxides in lipids and protecting membranes from peroxidation damage (14). GPX4 can be inactivated by intracellular glutathione (GSH), the synthesis of which depends on the import of extracellular cystine by system XC (cystine/glutamate transporter) that is composed of disulfide-linked heterodimers SLC7A11 and SLC3A2 (15). Studies have shown that ferroptosis plays a critical role in modulating various diseases, especially cancer (16,17). In gastric cancer, targeting genes that regulate ferroptosis possibly confer the drug resistance and repress cell growth (18,19). Thus, boosting ferroptosis is a plausible way to treat cancer.
Circular RNAs (circRNAs) are a new type of non-coding RNAs that exhibit poor protein coding ability (20). CircRNAs are generated by the back splicing of precursor messenger RNAs (mRNAs) and are characterized by a covalently closed structure, which makes them more stable in the physical environment than linear RNAs (21,22). High-throughput sequencing studies have revealed the abnormal expression profile of circRNAs in cancer tissues and cells (23,24). It has been suggested that circRNAs participate in multiple cellular processes, including cell growth, metastasis, and autophagy, by sponging microRNAs to modulate downstream mRNA expression or directly interacting with proteins (25). CircRHOT1 is a new circRNA that was identified in recent years and has been shown to promote cancer progression (26-29). Increasing evidence have identified the participation of circRHOT1 in proliferation, autophagy, apoptosis, and ferroptosis of various cancers such as hepatocellular carcinoma, breast cancer, pancreatic cancer (26,30,31). The diagnostic role of circRHOT1 has been identified in pancreatic cancer. Furthermore, the regulatory effects of circRHOT1 involve epigenetic regulation and post-transcriptional regulation. For example, circRHOT1 directly binds to miR-125a-3p, acting as an endogenous sponge to inhibit its activity, hence targets the E2F3 expression to promote proliferation and invasion of pancreatic cancer cells (32). In lung cancer, circRHOT1 modulate cell apoptosis and cell cycle via directly interact with acetyltransferase KAT5 to modulate the recruitment of RNA polymerase II and the histone H3 lysine 27 acetylation (H3k27Ac) modification to the promoter region of c-MYC (33). Therefore, targeting circRHOT1 is a promising strategy for cancer. However, the function of circRHOT1 in GC and ferroptosis is not yet clear. Based on the reported functions of circRHOT1 in cancer cell growth and death, we speculate that circRHOT1 may be a potential diagnostic factor and therapeutic target for GC.
In this study, we sought to explore the function of circRHOT1 in GC development. We determined GC cell ferroptosis by circRHOT1 depletion and identified GPX4 as a factor that mediates circRHOT1-regulated GC cell growth. Our findings suggest that circRHOT1 as an ideal target for inducing ferroptosis and could be used in the treatment of GC. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-550/rc).
MethodsOther Section
Clinical specimen
Tumor samples (n=50) and adjacent non-tumor tissues were collected from GC patients who were hospitalized and received surgery at the Qilu Hospital (Qingdao) from July 2018 to January 2022. All the samples were stored in liquid nitrogen and the expression of circRHOT1 was measured by quantitative real-time polymerase chain reaction (qRT-PCR). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from the patients before the study. The study involving human experiments was approved by the Medical Ethics Committee of the Qilu Hospital (Qingdao) (No. 2022-02-015).
Cell lines
The GC cell lines HGC-27 (CRL-5822), SGC-7901 (HTB-135), MKN-45 (CRL-5973), MGC-403 (CCL-230), and BGC-823 (CRL-1687) and GES-1 cell line (28200) were purchased from the Institute of Cells, ATCC (USA). All the cell lines were maintained in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum (Gibco, USA) and 1% penicillin and streptomycin (Sigma, USA) in a 37 ℃-humidified incubator filled with carbon dioxide.
qRT-PCR
Total RNA was extracted from the cells and tissues using Trizol reagent (Invitrogen, USA). The total RNAs were reverse-transcribed to complementary DNA using a first-strand synthesis kit (Thermo Fisher, USA). Gene expression was then calculated using SYBR (Synergy Brands) Green (Thermo Fisher, USA). Gene expression was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and U6 (snRNA U6), respectively.
Western blot
The tissues and cells were lysed with radioimmunoprecipitation assay lysis buffer (Thermo Fisher, USA). The cell lysates were divided in sodium dodecyl-sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. The protein bands were probed with primary antibodies against SLC7A11 (ab216876, Abcam), GPX4 (ab125066, Abcam), and β-actin (ab7817, Abcam) at 4 ℃ overnight, and then incubated with horseradish peroxidase-conjugated secondary antibodies. The protein bands were then visualized after reaction with enhanced chemiluminescence reagent (Millipore, Germany).
RNA FISH hybridization
The localization of circRHOT1 in the GC cells was detected using Cy3-labeled circRHOT1 probes that were synthesized by RiboBIo (China). The cells were incubated with the probes in accordance with the instructions of the fluorescence in situ hybridization (FISH) assay kit (RiboBio, China). The nuclei were stained with 4',6-diamidino-2-phenylindole (Sigma, USA). Images were taken under a confocal fluorescence microscope (Leica, Germany).
Cell treatment
Ectopic expression and gene silencing experiments were conducted to determine the effects of circRHOT1 and the molecular mechanism. The overexpression vectors of circRHOT1, KAT5, and GPX4, and shRNAs (i.e., shcircRNOT1, shKAT5, and shNC) were synthesized by RiboBio (China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen, USA) in accordance with the manufacturer’s instructions. Next, to induce ferroptosis, the cells were treated with a ferroptosis inducer erastin or inhibitor ferrostatin (5 µM; Selleck, USA) for 24 hours.
Cell Counting Kit 8 (CCK-8) assays
Cell viability was checked by CCK-8 assays (Beyotime, China) in accordance with the manufacturer’s instructions. In short, the cells were seeded into 96-well plates (5,000 cells/well) after the indicated treatment and incubated for 24, 48, 72, and 96 hours. CCK-8 reagent was then added to each well and hatched for 2 hours. The absorbance values at 450 nm were checked by a microplate reader (Thermo, USA).
Colony formation assays
To determine cell proliferation under alteration of circRHOT1, cell colony formation assay was conducted. The cells were transfected with indicated oligonucleotides, suspended as single cells, and seeded into a 6-well plate (1,000 cells/well). After incubation for 14 days, the visible colonies were stained with crystal violet for 20 minutes and captured with a microscope (Leica, Germany).
Lipid reactive oxygen species (ROS) detection
The level of lipid ROS was determined using the BODIOY C-11 probe (ABclonal, USA) in accordance with the manufacturer’s instructions. The cells were incubated with 5 µM probe for in the dark for 30 min at 37 ℃. Cells were then harvested and resuspended in serum-free medium. Samples were then analyzed by the BD FACs system.
Detection of iron, MDA, and GSH levels
The levels of intracellular iron, malondialdehyde (MDA), and GSH were measured by the iron assay kit (Abcam, USA), lipid peroxidation (MDA) assay kit (Sigma, USA), and GSH assay kit (BioVision, USA), respectively, in accordance with each manufacturer’s instructions. For MDA detection, cells were homogenized using MDA lysis buffer and centrifuged at 13,000 g for 3 min. After reaction with thiobarbituric acid (TBA), the absorbance values were measured at 532 nm. For GSH detection, cells were lysed with 5% 5-sulfosalicylic acid (SSA) solution and incubated with enzyme mix. Then absorbance values were detected at 450 nm.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed using the EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore, USA) in accordance with the manufacturer’s instructions. In short, the cells were fixed with 1% formaldehyde and lysed. The lysates were sonicated to obtain chromatin fragments around 500 bp. Antibodies against KAT5, H3k27Ac, RNA polymerase II, and IgG were incubated with the lysates overnight at 4 ℃. The next day, the lysates were hatched with protein A agarose beads. The precipitants were eluted, and RNA enrichment was analyzed by qRT-PCR and agarose gel electrophoresis.
Xenograft assays
The in vivo growth of cancer cells was determined by xenograft tumor model to further evaluate the effects of circRHOT1 on gastric cancer. A total of 15 healthy, male, SPF-grade (specific pathogen free), SCID (server combined immune-deficiency) nude mice, aged 6 weeks and weighing 15–20 g, were obtained from Vital River Laboratory (China). The mice were housed at a temperature of 20–24 ℃, with a relative humidity of 40–70%, on a 12-h light-dark cycle. Acclimatization was carried out for 7 days before the start of the formal experiment. The mice had 12-h light-dark and free access to food and water at all times during the experiment. The mice were randomized into shNC, shRHOT1, and shRHOT1 + KAT5 group with 5 in each group according to a Random number table method. Potential confounders were not considered. BGC-823 cells (5×106 cells per site) were inoculated into the fat pad of the SCID nude mice. All animals were included. Five days after inoculation, mice received intratumor injection of shNC, shRHOT1, and KAT5 overexpression vectors (20 nmol/20 g body weight) in 50 µL saline. The treatment was conducted every 3 days. The length and width of the tumors were measured every 5 days, and tumor size was calculated as follows: width2 × length/2. The mice were then sacrificed through cervical dislocation when the tumor size reached 1,000 mm3, and the tumors were collected for the following experiments.
The animal experiments were approved by the Ethics Committee of the Qilu Hospital (Qingdao) (No. KYDWLL-202327), in compliance with national guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Statistics
All the data were obtained from at least 3 independent experiments (technical replicates) and are presented as the mean ± standard deviation. The data analysis was performed using SPSS 20.0 software and GraphPad Prism 7 (Public Version). For analyses of datasets with parametric distribution, the two-tailed Student’s t-test and a one-way analysis of variance followed by pair-wise comparisons were used for the comparisons between two or multiple groups. For the analysis of datasets with non-parametric distribution, the differences between two or more groups were compared using Mann-Whitney U test and Kruskal-Wallis test with Dunn’s multiple comparisons post-test, respectively. P values <0.05 were considered statistically significant.
ResultsOther Section
CircRHOT1 expression was upregulated in the GC tissues
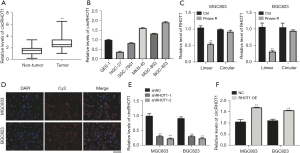
We first investigated the expression of circRHOT1 in GC. As Figure 1A shows, circRHOT1 was significantly overexpressed in the GC tumor tissues compared to the non-tumor adjacent tissues. To elucidate the function of circRHOT1 in GC, we evaluated the expression of circRHOT1 in several GC cell lines and selected the MGC803 and BGC823 cell lines, which expressed high levels of circRHOT1 (Figure 1B), for a mechanism study. We observed that treatment with Rnase (Ribonuclease) R significantly suppressed the level of the linear RHOT1 rather than the circular RHOT1 (Figure 1C) in the GC cells. Further, the FISH experiment showed that Cy3 (Cyanine 3)-labeled circRHOT1 was located in the cytoplasm of the GC cells (Figure 1D). Transfection with the shRNAs and overexpression vectors that target circRHOT1 significantly suppressed or elevated the RNA levels of circRHOT1 (Figure 1E,1F).
CircRHOT1 suppression induced ferroptosis in the GC cells in vitro
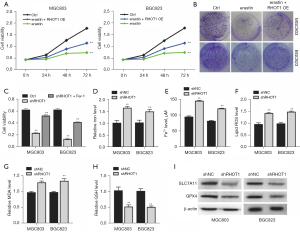
Erastin is an inducer of ferroptosis. We found that the overexpression of circRHOT1 led to the recovery of cell growth, which had been suppressed by the erastin-induced ferroptosis in the GC cells (Figure 2A,2B). Moreover, the knockdown of circRHOT1 suppressed GC cell viability, which was recovered by the ferroptosis inhibitor ferrostatin (Fer-1) (Figure 2C). These results indicated that circRHOT1 may promote GC cell proliferation by suppressing ferroptosis. We next examined the features of ferroptosis. The knockdown of circRHOT1 significantly elevated the accumulation of total iron, Fe2+, lipid ROS, and MDA, and decreased the level of GSH in the GC cells (Figure 2D-2H), indicating the generation of ferroptosis. Additionally, circRHOT1 depletion downregulated the expression of SLC7A11 and GPX4 (Figure 2I), the representative ferroptosis suppressors, in the GC cells.
CircRHOT1 epigenetically modulated GPX4 expression in the GC cells by recruiting KAT5 in vitro
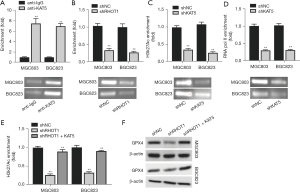
Previous studies have shown that circRHOT1 recruits histone acetyltransferase KAT5 to epigenetically modulate gene expression (30,33). The ChIP assay results showed that KAT5 interacted with the GPX4 gene in the GC cells (Figure 3A). The knockdown of circRHOT1 significantly decreased the enrichment of KAT5 in the GPX4 gene (Figure 3B). Moreover, the depletion of KAT5 significantly suppressed the acetylation of lysine 27 on histone H3 protein subunit (H3k27Ac) and the enrichment of RNA polymerase II in the GPX4 gene (Figure 3C,3D). The overexpression of KAT5 recovered the shRHOT1-decreased H3k27Ac of GPX4 gene (Figure 3E), and elevated GPX4 expression (Figure 3F). These data suggested that KAT5 may mediate circRHOT1-modulated GPX4 expression.
Restoration of KAT5 and GPX4 rescued circRHOT1 deletion-induced GC cell death in vitro
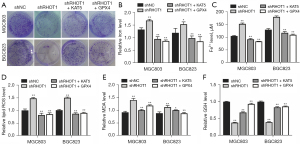
To verify the function of KAT5 and GPX4 in circRHOT1-mediated GC cell growth, we performed a knockdown and rescue experiment. The colony formation results showed that the suppressed colony formation ability upon circRHOT1 depletion was reversed by the overexpression of KAT5 and GPX4 (Figure 4A). Further, the ferroptosis biomarkers indicated that circRHOT1 depletion caused the accumulation of total iron, Fe2+, lipid ROS, and MDA, and decreased the level of GSH in GC cells, which were recovered by the overexpression of KAT5 and GPX4 (Figure 4B-4F).
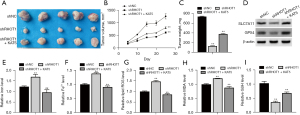
Restoration of KAT5 impairs circRHOT1 deletion-suppressed GC tumor growth in vivo
We then confirmed the circRHOT1/KAT5/GPX4 axis in an in vivo model. We observed that the knockdown of circRHOT1 significantly suppressed GC tumor growth in the mouse model, whereas KAT5 overexpression reduced this effect (Figure 5A-5C). Consistent with the results of the in vitro experiments, KAT5 overexpression also upregulated the protein levels of SLC7A11 and GPX4 that had been suppressed by circRHOT1 depletion in the GC tumors (Figure 5D). The accumulated levels of iron, Fe2+, lipid ROS, and MDA, and the decreased the level of GSH in the circRHOT1 depletion group were reversed by KAT5 overexpression (Figure 5E-5I).
DiscussionOther Section
In recent years, the functions of non-coding RNAs, especially circRNAs, have drawn great attention (25). The involvement of circRNAs in human diseases, especially cancers, have been widely reported (20). In this study, we found that the expression of circRHOT1 was elevated in the tumor tissues of the GC patients and the knockdown of circRHOT1 promoted the ferroptosis of the GC cells. Ferroptosis is a unique form of lipid peroxidation-induced cell death, which is characterized by the excess accumulation of intracellular iron and lipid ROS, and a decreased level of the intracellular antioxidation enzyme GSH (34). We observed that the overexpression of circRHOT1 suppressed erastin-induced ferroptosis, and the knockdown of circRHOT1 stimulated ferroptosis, and altered the expression of SLC7A11 and GPX4, a well-recognized ferroptosis suppressor.
Studies have suggested that circRHOT1 is a potential diagnostic biomarker and therapeutic target for cancers. For example, circRHOT1 expression was found to be elevated in pancreatic cancer cell lines and tissues, and to directly bind to miR-125a-3p as an endogenous sponge to impede its targeting activity (32). The depletion of circRHOT1 was shown to dramatically suppress pancreatic cancer cell proliferation and invasion and enhance cell apoptosis by regulating the downstream E2F3 function (26). In breast cancer cells, circRHOT1 acts as a sponge of miR-106a-5p and suppresses the ferroptosis of cancer cells by targeting signal transducer and activator of transcription 3 (31). Other than sponging miRNAs, circRHOT1 also modulates gene expression by epigenetically regulating gene transcription. It has been reported that circRHOT1 recruits KAT5 to the promoter region of the NR2F6 gene and activates NR2F6 transcription, which consequently promotes the cell growth, migration, and invasion of hepatocellular carcinoma cells (30). The knockdown of circRHOT1 has been shown to downregulate the transcription of c-MYC and suppresses the proliferation of non-small cell lung cancer cells (33). In this study, we showed that circRHOT1 recruited KAT5 to the promoter region of GPX4 and triggered its expression in the GC cells.
KAT5, also known as Tip60, is a widely reported member of the histone acetyltransferase complexes that epigenetically modulate gene transcription via the enrichment of transcription active marker histone H3 lysine 27 acetylation (H3k27Ac) in the promoter regions of target genes and trigger gene transcription (35). Numerous studies have shown the important role of KAT5 in cancer progression and its potential as a cancer therapeutic target (36,37). The overexpression of KAT5 has been shown to be closely correlated with the metastasis of anaplastic thyroid cancer and to stabilize the c-MYC to facilitate cancer cell proliferation and invasion (38). The modifications of KAT5, such as phosphorylation and O-GlcNAcylation, are also reported to be involved in cancer development and metastasis (39,40). We found that KAT5 directly interacted with the GPX4 gene, and KAT5 depletion suppressed the enrichment of H3k27Ac and RNA polymerase II on the promoter of GPX4. The knockdown of circRHOT1 also led to suppressed H3k27Ac, whereas the overexpression of KAT5 reduced this effect. GPX4 is a well-known suppressor and pivotal regulator of ferroptosis and could remove the hydrogen peroxide products from membrane lipids and prevent intracellular oxidative stress (14). Here, we found that KAT5 could epigenetically regulate the acetylation and expression of GPX4 in GC cells, and circRHOT1 promoted the recruitment of KAT5 to GPX4, suggesting that circRHOT1 may suppress ferroptosis via GPX4. However, further studies are needed to confirm the perspective, such as in situ tumor model, and other potential downstream signaling of circRHOT3 in GC and ferroptosis should be explored in future study.
ConclusionsOther Section
To summarize, circRHOT1 was overexpressed in the GC tumor tissues compared to the non-tumor adjacent tissues. The overexpression of circRHOT1 suppressed ferroptosis in the GC cells by recruiting KAT5 to epigenetically promote GPX4 expression and function. This work supports that developing therapeutics targeting circRHOT1 in GC is a promising option for GC patients.
AcknowledgmentsOther Section
Funding: None.
FootnoteOther Section
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-550/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-550/dss
Peer Review File: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-550/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-550/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study involving human experiments was approved by the Medical Ethics Committee of the Qilu Hospital (Qingdao) (No. 2022-02-015). Informed consent was taken from all the patients. The animal experiments were approved by the Ethics Committee of the Qilu Hospital (Qingdao) (No. KYDWLL-202327), in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Johnston FM, Beckman M. Updates on Management of Gastric Cancer. Curr Oncol Rep 2019;21:67. [Crossref] [PubMed]
- De Re V. Molecular Features Distinguish Gastric Cancer Subtypes. Int J Mol Sci 2018;19:3121. [Crossref] [PubMed]
- Venerito M, Link A, Rokkas T, et al. Gastric cancer - clinical and epidemiological aspects. Helicobacter 2016;21:39-44. [Crossref] [PubMed]
- Patel TH, Cecchini M. Targeted Therapies in Advanced Gastric Cancer. Curr Treat Options Oncol 2020;21:70. [Crossref] [PubMed]
- Xiao S, Zhou L. Gastric cancer: Metabolic and metabolomics perspectives Int J Oncol 2017;51:5-17. (Review). [Crossref] [PubMed]
- Ucaryilmaz Metin C, Ozcan G. Comprehensive bioinformatic analysis reveals a cancer-associated fibroblast gene signature as a poor prognostic factor and potential therapeutic target in gastric cancer. BMC Cancer 2022;22:692. [Crossref] [PubMed]
- Tan Z. Recent Advances in the Surgical Treatment of Advanced Gastric Cancer: A Review. Med Sci Monit 2019;25:3537-41. [Crossref] [PubMed]
- Sexton RE, Al Hallak MN, Diab M, et al. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev 2020;39:1179-203. [Crossref] [PubMed]
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet 2020;396:635-48. [Crossref] [PubMed]
- Battaglia AM, Chirillo R, Aversa I, et al. Ferroptosis and Cancer: Mitochondria Meet the "Iron Maiden" Cell Death. Cells 2020;9:1505. [Crossref] [PubMed]
- Li D, Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct Target Ther 2020;5:108. [Crossref] [PubMed]
- Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 2019;12:34. [Crossref] [PubMed]
- Wang Y, Wei Z, Pan K, et al. The function and mechanism of ferroptosis in cancer. Apoptosis 2020;25:786-98. [Crossref] [PubMed]
- Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell 2019;35:830-49. [Crossref] [PubMed]
- Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol 2021;18:280-96. [Crossref] [PubMed]
- Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer 2022;22:381-96. [Crossref] [PubMed]
- Jiang M, Hu R, Yu R, et al. A narrative review of mechanisms of ferroptosis in cancer: new challenges and opportunities. Ann Transl Med 2021;9:1599. [Crossref] [PubMed]
- Geng D, Wu H. Abrogation of ARF6 in promoting erastin-induced ferroptosis and mitigating capecitabine resistance in gastric cancer cells. J Gastrointest Oncol 2022;13:958-67. [Crossref] [PubMed]
- Weng W, Zhang M, Ni S, et al. Decreased expression of claudin-18.2 in alpha-fetoprotein-producing gastric cancer compared to conventional gastric cancer. J Gastrointest Oncol 2022;13:1035-45. [Crossref] [PubMed]
- Li R, Jiang J, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci 2020;77:1661-80. [Crossref] [PubMed]
- Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol 2019;12:90. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer 2020;19:30. [Crossref] [PubMed]
- Xu X, Zhang J, Tian Y, et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer 2020;19:128. [Crossref] [PubMed]
- Chen L, Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett 2021;505:49-57. [Crossref] [PubMed]
- Qu S, Hao X, Song W, et al. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics 2019;11:53-63. [Crossref] [PubMed]
- Wang Y, Li Z, Xu S, et al. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J Clin Lab Anal 2020;34:e23359. [Crossref] [PubMed]
- Zhang Q, Cheng F, Zhang Z, et al. Propofol suppresses non-small cell lung cancer tumorigenesis by regulation of circ-RHOT1/miR-326/FOXM1 axis. Life Sci 2021; Epub ahead of print. [Crossref]
- Ke H, Zhang J, Wang F, et al. ZNF652-Induced circRHOT1 Promotes SMAD5 Expression to Modulate Tumorigenic Properties and Nature Killer Cell-Mediated Toxicity in Bladder Cancer via Targeting miR-3666. J Immunol Res 2021;2021:7608178. [Crossref] [PubMed]
- Wang L, Long H, Zheng Q, et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer 2019;18:119. [Crossref] [PubMed]
- Zhang H, Ge Z, Wang Z, et al. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging (Albany NY) 2021;13:8115-26. [Crossref] [PubMed]
- Ling S, He Y, Li X, et al. CircRHOT1 mediated cell proliferation, apoptosis and invasion of pancreatic cancer cells by sponging miR-125a-3p. J Cell Mol Med 2020;24:9881-9. [Crossref] [PubMed]
- Ren X, Yu J, Guo L, et al. Circular RNA circRHOT1 contributes to pathogenesis of non-small cell lung cancer by epigenetically enhancing C-MYC expression through recruiting KAT5. Aging (Albany NY) 2021;13:20372-82. [Crossref] [PubMed]
- Liang C, Zhang X, Yang M, et al. Recent Progress in Ferroptosis Inducers for Cancer Therapy. Adv Mater 2019;31:e1904197. [Crossref] [PubMed]
- Cregan S, McDonagh L, Gao Y, et al. KAT5 (Tip60) is a potential therapeutic target in malignant pleural mesothelioma. Int J Oncol 2016;48:1290-6. [Crossref] [PubMed]
- Du J, Fu L, Ji F, et al. FosB recruits KAT5 to potentiate the growth and metastasis of papillary thyroid cancer in a DPP4-dependent manner. Life Sci 2020;259:118374. [Crossref] [PubMed]
- McGuire A, Casey MC, Shalaby A, et al. Quantifying Tip60 (Kat5) stratifies breast cancer. Sci Rep 2019;9:3819. [Crossref] [PubMed]
- Wei X, Cai S, Boohaker RJ, et al. KAT5 promotes invasion and metastasis through C-MYC stabilization in ATC. Endocr Relat Cancer 2019;26:141-51. [Crossref] [PubMed]
- Liu R, Gou D, Xiang J, et al. O-GlcNAc modified-TIP60/KAT5 is required for PCK1 deficiency-induced HCC metastasis. Oncogene 2021;40:6707-19. [Crossref] [PubMed]
- Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature 2013;498:70-4. [Crossref] [PubMed]


